Coxsackievirus A16 in Southern Vietnam
- PMID: 34248913
- PMCID: PMC8265502
- DOI: 10.3389/fmicb.2021.689658
Coxsackievirus A16 in Southern Vietnam
Abstract
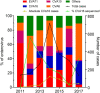
Background: Hand, Foot and Mouth Disease (HFMD) is a major public health concern in the Asia-Pacific region. Most recent HFMD outbreaks have been caused by enterovirus A71 (EV-A71), coxsackievirus A16 (CVA16), CVA10, and CVA6. There has been no report regarding the epidemiology and genetic diversity of CVA16 in Vietnam. Such knowledge is critical to inform the development of intervention strategies. Materials and Methods: From 2011 to 2017, clinical samples were collected from in- and outpatients enrolled in a HFMD research program conducted at three referral hospitals in Ho Chi Minh City (HCMC), Vietnam. Throat or rectal swabs positive for CVA16 with sufficient viral load were selected for whole genome sequencing and evolutionary analysis. Results: Throughout the study period, 320 CVA16 positive samples were collected from 2808 HFMD patients (11.4%). 59.4% of patients were male. The median age was 20.8 months (IQR, 14.96-31.41). Patients resided in HCMC (55.3%), Mekong Delta (22.2%), and South East Vietnam (22.5%). 10% of CVA16 infected patients had moderately severe or severe HFMD. CVA16 positive samples from 153 patients were selected for whole genome sequencing, and 66 complete genomes were obtained. Phylogenetic analysis demonstrated that Vietnamese CVA16 strains belong to a single genogroup B1a that clusters together with isolates from China, Japan, Thailand, Malaysia, France and Australia. The CVA16 strains of the present study were circulating in Vietnam some 4 years prior to its detection in HFMD cases. Conclusion: We report for the first time on the molecular epidemiology of CVA16 in Vietnam. Unlike EV-A71, which showed frequent replacement between subgenogroups B5 and C4 every 2-3 years in Vietnam, CVA16 displays a less pronounced genetic alternation with only subgenogroup B1a circulating in Vietnam since 2011. Our collective findings emphasize the importance of active surveillance for viral circulation in HFMD endemic countries, critical to informing outbreak response and vaccine development.
Keywords: Vietnam; coxsackievirus A16; evolution; hand foot mouth disease; picornavirus.
Copyright © 2021 Nhu, Nhan, Anh, Hong, Van, Thanh, Hang, Han, Ny, Nguyet, Quy, Qui, Khanh, Hung, Tuan, Chau, Thwaites, Doorn and Tan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

