In vitro Characterization of Fitness and Convalescent Antibody Neutralization of SARS-CoV-2 Cluster 5 Variant Emerging in Mink at Danish Farms
- PMID: 34248922
- PMCID: PMC8267889
- DOI: 10.3389/fmicb.2021.698944
In vitro Characterization of Fitness and Convalescent Antibody Neutralization of SARS-CoV-2 Cluster 5 Variant Emerging in Mink at Danish Farms
Abstract
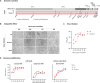
In addition to humans, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can transmit to animals that include hamsters, cats, dogs, mink, ferrets, tigers, lions, cynomolgus macaques, rhesus macaques, and treeshrew. Among these, mink are particularly susceptible. Indeed, 10 countries in Europe and North America reported SARS-CoV-2 infection among mink on fur farms. In Denmark, SARS-CoV-2 spread rapidly among mink farms and spilled-over back into humans, acquiring mutations/deletions with unknown consequences for virulence and antigenicity. Here we describe a mink-associated SARS-CoV-2 variant (Cluster 5) characterized by 11 amino acid substitutions and four amino acid deletions relative to Wuhan-Hu-1. Temporal virus titration, together with genomic and subgenomic viral RNA quantitation, demonstrated a modest in vitro fitness attenuation of the Cluster 5 virus in the Vero-E6 cell line. Potential alterations in antigenicity conferred by amino acid changes in the spike protein that include three substitutions (Y453F, I692V, and M1229I) and a loss of two amino acid residues 69 and 70 (ΔH69/V70), were evaluated in a virus microneutralization assay. Compared to a reference strain, the Cluster 5 variant showed reduced neutralization in a proportion of convalescent human COVID-19 samples. The findings underscore the need for active surveillance SARS-CoV-2 infection and virus evolution in susceptible animal hosts.
Keywords: COVID-19; SARS-CoV-2; coronavirus; mink; virus neutralization.
Copyright © 2021 Lassaunière, Fonager, Rasmussen, Frische, Polacek, Rasmussen, Lohse, Belsham, Underwood, Winckelmann, Bollerup, Bukh, Weis, Sækmose, Aagaard, Alfaro-Núñez, Mølbak, Bøtner and Fomsgaard.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

