Comparative analysis of fatty acid metabolism based on transcriptome sequencing of wild and cultivated Ophiocordyceps sinensis
- PMID: 34249512
- PMCID: PMC8255070
- DOI: 10.7717/peerj.11681
Comparative analysis of fatty acid metabolism based on transcriptome sequencing of wild and cultivated Ophiocordyceps sinensis
Abstract
Background: Ophiocordyceps sinensis is a species endemic to the alpine and high-altitude areas of the Qinghai-Tibet plateau. Although O. sinensis has been cultivated since the past few years, whether cultivated O. sinensis can completely replace wild O. sinensis remains to be determined.
Methods: To explore the differences of O. sinensis grown in varied environments, we conducted morphological and transcriptomic comparisons between wild and cultivated samples who with the same genetic background.
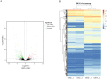
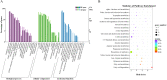
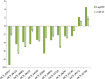
Results: The results of morphological anatomy showed that there were significant differences between wild and cultivated O. sinensis, which were caused by different growth environments. Then, a total of 9,360 transcripts were identified using Illumina paired-end sequencing. Differential expression analysis revealed that 73.89% differentially expressed genes (DEGs) were upregulated in O. sinensis grown under natural conditions compared with that grown under artificial conditions. Functional enrichment analysis showed that some key DEGs related to fatty acid metabolism, including acyl-CoA dehydrogenase, enoyl-CoA hydratase, 3-ketoacyl-CoA thiolase, and acetyl-CoA acetyltransferase, were upregulated in wild O. sinensis. Furthermore, gas chromatography-mass spectrometry results confirmed that the fatty acid content of wild O. sinensis was significantly higher than that of cultivated O. sinensis and that unsaturated fatty acids accounted for a larger proportion.
Conclusion: These results provide a theoretical insight to the molecular regulation mechanism that causes differences between wild and cultivated O. sinensis and improving artificial breeding.
Keywords: Cultivated; Fatty acid metabolism; GC-MS; Ophiocordyceps sinensis; Transcriptome; Wild-grown.
©2021 Zhang et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


Similar articles
-
Integrated metabolomics and transcriptomics reveal metabolites difference between wild and cultivated Ophiocordyceps sinensis.Food Res Int. 2023 Jan;163:112275. doi: 10.1016/j.foodres.2022.112275. Epub 2022 Nov 29. Food Res Int. 2023. PMID: 36596185
-
Discovery of differential sequences for improving breeding and yield of cultivated Ophiocordyceps sinensis through ITS sequencing and phylogenetic analysis.Chin J Nat Med. 2018 Oct;16(10):749-755. doi: 10.1016/S1875-5364(18)30114-6. Chin J Nat Med. 2018. PMID: 30322608
-
Morphological Observations and Fatty Acid Composition of Indoor-Cultivated Cordyceps sinensis at a High-Altitude Laboratory on Sejila Mountain, Tibet.PLoS One. 2015 May 4;10(5):e0126095. doi: 10.1371/journal.pone.0126095. eCollection 2015. PLoS One. 2015. PMID: 25938484 Free PMC article.
-
Comparative transcriptome analysis revealed genes involved in the fruiting body development of Ophiocordyceps sinensis.PeerJ. 2020 Jan 16;8:e8379. doi: 10.7717/peerj.8379. eCollection 2020. PeerJ. 2020. PMID: 31988806 Free PMC article.
-
Transcriptome analysis reveals the molecular mechanism of hepatic fat metabolism disorder caused by Muscovy duck reovirus infection.Avian Pathol. 2018 Apr;47(2):127-139. doi: 10.1080/03079457.2017.1380294. Epub 2017 Oct 10. Avian Pathol. 2018. PMID: 28911249 Clinical Trial.
Cited by
-
Changes in the content of pollen total lipid and TAG in Arabidopsis thaliana DGAT1 mutant as11.AoB Plants. 2023 Mar 22;15(2):plad012. doi: 10.1093/aobpla/plad012. eCollection 2023 Feb. AoB Plants. 2023. PMID: 37064996 Free PMC article.
-
Evaluation of Cordyceps sinensis Quality in 15 Production Areas Using Metabolomics and the Membership Function Method.J Fungi (Basel). 2024 May 16;10(5):356. doi: 10.3390/jof10050356. J Fungi (Basel). 2024. PMID: 38786711 Free PMC article.
-
Upcycling Chitin Waste and Aged Rice into Fungi Protein Through Fermentation with Cordyceps militaris.J Fungi (Basel). 2025 Apr 16;11(4):315. doi: 10.3390/jof11040315. J Fungi (Basel). 2025. PMID: 40278135 Free PMC article.
-
Effect of Air Drying on the Metabolic Profile of Fresh Wild and Artificial Cordyceps sinensis.Foods. 2023 Dec 21;13(1):48. doi: 10.3390/foods13010048. Foods. 2023. PMID: 38201076 Free PMC article.
-
Evaluation of Lipid Changes During the Drying Process of Cordyceps sinensis by Ultra Performance Liquid Chromatography-Tandem Mass Spectrometry (UPLC-MS/MS)-Based Lipidomics Technique.J Fungi (Basel). 2024 Dec 11;10(12):855. doi: 10.3390/jof10120855. J Fungi (Basel). 2024. PMID: 39728352 Free PMC article.
References
-
- Dong CH, Wen-Jia LI, Zeng-Zhi LI, Yan WJ, Tai-Hui LI, Liu XZ. Cordyceps industry in China: current status, challenges and perspectives—Jinhu declaration for cordyceps industry development. Mycosystema. 2016;35:1–15.
-
- Dong-Wei X, Xiao-Nan W, Lian-Shuang FU, Jian S, Tao G, Zhuo-Fu LI. Effects of low temperature stress on membrane fatty acid in tillering node of winter wheat. Journal of Triticeae Crops. 2013;33:746–751.
LinkOut - more resources
Full Text Sources
Miscellaneous

