Simplified Epigenome Profiling Using Antibody-tethered Tagmentation
- PMID: 34250209
- PMCID: PMC8250384
- DOI: 10.21769/BioProtoc.4043
Simplified Epigenome Profiling Using Antibody-tethered Tagmentation
Abstract
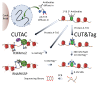
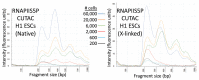
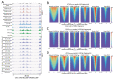
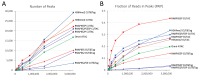
We previously introduced Cleavage Under Targets & Tagmentation (CUT&Tag), an epigenomic profiling method in which antibody tethering of the Tn5 transposase to a chromatin epitope of interest maps specific chromatin features in small samples and single cells. With CUT&Tag, intact cells or nuclei are permeabilized, followed by successive addition of a primary antibody, a secondary antibody, and a chimeric Protein A-Transposase fusion protein that binds to the antibody. Addition of Mg++ activates the transposase and inserts sequencing adapters into adjacent DNA in situ. We have since adapted CUT&Tag to also map chromatin accessibility by simply modifying the transposase activation conditions when using histone H3K4me2, H3K4me3, or Serine-5-phosphorylated RNA Polymerase II antibodies. Using these antibodies, we redirect the tagmentation of accessible DNA sites to produce chromatin accessibility maps with exceptionally high signal-to-noise and resolution. All steps from nuclei to amplified sequencing-ready libraries are performed in single PCR tubes using non-toxic reagents and inexpensive equipment, making our simplified strategy for simultaneous chromatin profiling and accessibility mapping suitable for the lab, home workbench, or classroom.
Keywords: CUT&Tag; Chromatin accessibility; Epigenomic profiling; Histone modifications; RNA polymerase II.
©Copyright Henikoff et al.
Conflict of interest statement
Competing interestsSH has filed patent applications related to this work. JGH and KA declare no competing interests.
Figures

References
-
- Andersson R., Sandelin A. and Danko C. G.(2015). A unified architecture of transcriptional regulatory elements. Trends Genet 31(8): 426-433. - PubMed
-
- Bownes M.(1990). Preferential insertion of P elements into genes expressed in the germ-line of Drosophila melanogaster . Mol Gen Genet 222(2-3): 457-460. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

