Repeated evolution of circadian clock dysregulation in cavefish populations
- PMID: 34252077
- PMCID: PMC8297936
- DOI: 10.1371/journal.pgen.1009642
Repeated evolution of circadian clock dysregulation in cavefish populations
Abstract
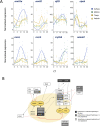
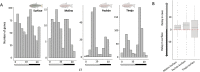
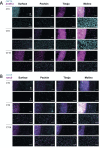
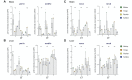
Circadian rhythms are nearly ubiquitous throughout nature, suggesting they are critical for survival in diverse environments. Organisms inhabiting largely arrhythmic environments, such as caves, offer a unique opportunity to study the evolution of circadian rhythms in response to changing ecological pressures. Populations of the Mexican tetra, Astyanax mexicanus, have repeatedly invaded caves from surface rivers, where individuals must contend with perpetual darkness, reduced food availability, and limited fluctuations in daily environmental cues. To investigate the molecular basis for evolved changes in circadian rhythms, we investigated rhythmic transcription across multiple independently-evolved cavefish populations. Our findings reveal that evolution in a cave environment has led to the repeated disruption of the endogenous biological clock, and its entrainment by light. The circadian transcriptome shows widespread reductions and losses of rhythmic transcription and changes to the timing of the activation/repression of core-transcriptional clock. In addition to dysregulation of the core clock, we find that rhythmic transcription of the melatonin regulator aanat2 and melatonin rhythms are disrupted in cavefish under darkness. Mutants of aanat2 and core clock gene rorca disrupt diurnal regulation of sleep in A. mexicanus, phenocopying circadian modulation of sleep and activity phenotypes of cave populations. Together, these findings reveal multiple independent mechanisms for loss of circadian rhythms in cavefish populations and provide a platform for studying how evolved changes in the biological clock can contribute to variation in sleep and circadian behavior.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

