Assessing sensitivity and reproducibility of RT-ddPCR and RT-qPCR for the quantification of SARS-CoV-2 in wastewater
- PMID: 34252511
- PMCID: PMC8267102
- DOI: 10.1016/j.jviromet.2021.114230
Assessing sensitivity and reproducibility of RT-ddPCR and RT-qPCR for the quantification of SARS-CoV-2 in wastewater
Abstract
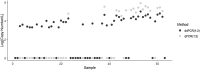
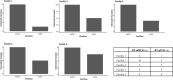
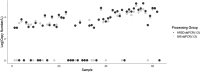
Throughout the COVID-19 global pandemic there has been significant interest and investment in using Wastewater-Based Epidemiology (WBE) for surveillance of viral pathogen presence and infections at the community level. There has been a push for widescale implementation of standardized protocols to quantify viral loads in a range of wastewater systems. To address concerns regarding sensitivity, limits of quantification, and large-scale reproducibility, a comparison of two similar workflows using RT-qPCR and RT-ddPCR was conducted. Sixty raw wastewater influent samples were acquired from nine distinct wastewater treatment plants (WWTP's) served by the Hampton Roads Sanitation District (HRSD, Virginia Beach, Virginia) over a 6-month period beginning March 9th, 2020. Common reagents, controls, master mixes and nucleic acid extracts were shared between two individual processing groups based out of HRSD and the UNC Chapel Hill Institute of Marine Sciences (IMS, Morehead City, North Carolina). Samples were analyzed in parallel using One-Step RT-qPCR and One-Step RT-ddPCR with Nucleocapsid Protein 2 (N2) specific primers and probe. Influent SARS-CoV-2 N2 concentrations steadily increased over time spanning a range from non-detectable to 2.13E + 05 copies/L. Systematic dilution of the extracts indicated that inhibitory components in the wastewater matrices did not significantly impede the detection of a positive N2 signal for either workflow. The RT-ddPCR workflow had a greater analytical sensitivity with a lower Limit of Detection (LOD) at 0.066 copies/μl of template compared to RT-qPCR with a calculated LOD of 12.0 copies/μL of template. Interlaboratory comparisons using non-parametric correlation analysis demonstrated that there was a strong, significant, positive correlation between split extracts when employing RT-ddPCR for analysis with a ρ value of 0.86.
Keywords: Molecular methods; PCR; SARS-CoV-2; Wastewater.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors report no declarations of interest.
Figures

Similar articles
-
Tracking the temporal variation of COVID-19 surges through wastewater-based epidemiology during the peak of the pandemic: A six-month long study in Charlotte, North Carolina.Sci Total Environ. 2022 Mar 25;814:152503. doi: 10.1016/j.scitotenv.2021.152503. Epub 2021 Dec 23. Sci Total Environ. 2022. PMID: 34954186 Free PMC article.
-
Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance.Sci Total Environ. 2022 Jan 20;805:149877. doi: 10.1016/j.scitotenv.2021.149877. Epub 2021 Aug 25. Sci Total Environ. 2022. PMID: 34818780 Free PMC article. Review.
-
Comparison of Different Reverse Transcriptase-Polymerase Chain Reaction-Based Methods for Wastewater Surveillance of SARS-CoV-2: Exploratory Study.JMIR Public Health Surveill. 2024 Aug 19;10:e53175. doi: 10.2196/53175. JMIR Public Health Surveill. 2024. PMID: 39158943 Free PMC article.
-
Analytical and Clinical Performance of Droplet Digital PCR in the Detection and Quantification of SARS-CoV-2.Mol Diagn Ther. 2021 Sep;25(5):617-628. doi: 10.1007/s40291-021-00547-1. Epub 2021 Jul 28. Mol Diagn Ther. 2021. PMID: 34319580 Free PMC article.
-
Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance.Water Res. 2021 Sep 15;203:117516. doi: 10.1016/j.watres.2021.117516. Epub 2021 Aug 5. Water Res. 2021. PMID: 34412018 Free PMC article. Review.
Cited by
-
Assessment of two volumetrically different concentration approaches to improve sensitivities for SARS-CoV-2 detection during wastewater monitoring.J Virol Methods. 2023 Jan;311:114645. doi: 10.1016/j.jviromet.2022.114645. Epub 2022 Nov 1. J Virol Methods. 2023. PMID: 36332716 Free PMC article.
-
Tracking the temporal variation of COVID-19 surges through wastewater-based epidemiology during the peak of the pandemic: A six-month long study in Charlotte, North Carolina.Sci Total Environ. 2022 Mar 25;814:152503. doi: 10.1016/j.scitotenv.2021.152503. Epub 2021 Dec 23. Sci Total Environ. 2022. PMID: 34954186 Free PMC article.
-
Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance.Sci Total Environ. 2022 Jan 20;805:149877. doi: 10.1016/j.scitotenv.2021.149877. Epub 2021 Aug 25. Sci Total Environ. 2022. PMID: 34818780 Free PMC article. Review.
-
Wastewater surveillance demonstrates high predictive value for COVID-19 infection on board repatriation flights to Australia.Environ Int. 2022 Jan;158:106938. doi: 10.1016/j.envint.2021.106938. Epub 2021 Oct 14. Environ Int. 2022. PMID: 34735954 Free PMC article.
-
A scoping review of global SARS-CoV-2 wastewater-based epidemiology in light of COVID-19 pandemic.Heliyon. 2024 May 3;10(9):e30600. doi: 10.1016/j.heliyon.2024.e30600. eCollection 2024 May 15. Heliyon. 2024. PMID: 38765075 Free PMC article.
References
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. - DOI - PMC - PubMed
-
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., et al. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

