Sequence-specific minimizers via polar sets
- PMID: 34252928
- PMCID: PMC8686682
- DOI: 10.1093/bioinformatics/btab313
Sequence-specific minimizers via polar sets
Abstract
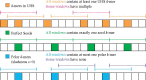
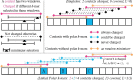
Motivation: Minimizers are efficient methods to sample k-mers from genomic sequences that unconditionally preserve sufficiently long matches between sequences. Well-established methods to construct efficient minimizers focus on sampling fewer k-mers on a random sequence and use universal hitting sets (sets of k-mers that appear frequently enough) to upper bound the sketch size. In contrast, the problem of sequence-specific minimizers, which is to construct efficient minimizers to sample fewer k-mers on a specific sequence such as the reference genome, is less studied. Currently, the theoretical understanding of this problem is lacking, and existing methods do not specialize well to sketch specific sequences.
Results: We propose the concept of polar sets, complementary to the existing idea of universal hitting sets. Polar sets are k-mer sets that are spread out enough on the reference, and provably specialize well to specific sequences. Link energy measures how well spread out a polar set is, and with it, the sketch size can be bounded from above and below in a theoretically sound way. This allows for direct optimization of sketch size. We propose efficient heuristics to construct polar sets, and via experiments on the human reference genome, show their practical superiority in designing efficient sequence-specific minimizers.
Availability and implementation: A reference implementation and code for analyses under an open-source license are at https://github.com/kingsford-group/polarset.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Blackburn S.R. (2015) Non-overlapping codes. IEEE Trans. Inf. Theory, 61, 4890–4894.
-
- DeBlasio D. et al. (2019) Practical universal k-mer sets for minimizer schemes. In Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, BCB ’19, ACM, New York, NY, USA. pp. 167–176.
-
- Deorowicz S. et al. (2015) KMC 2: fast and resource-frugal k-mer counting. Bioinformatics, 31, 1569–1576. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

