Asynchronous parallel Bayesian optimization for AI-driven cloud laboratories
- PMID: 34252975
- PMCID: PMC8275326
- DOI: 10.1093/bioinformatics/btab291
Asynchronous parallel Bayesian optimization for AI-driven cloud laboratories
Abstract
Motivation: The recent emergence of cloud laboratories-collections of automated wet-lab instruments that are accessed remotely, presents new opportunities to apply Artificial Intelligence and Machine Learning in scientific research. Among these is the challenge of automating the process of optimizing experimental protocols to maximize data quality.
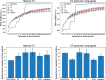
Results: We introduce a new deterministic algorithm, called PaRallel OptimizaTiOn for ClOud Laboratories (PROTOCOL), that improves experimental protocols via asynchronous, parallel Bayesian optimization. The algorithm achieves exponential convergence with respect to simple regret. We demonstrate PROTOCOL in both simulated and real-world cloud labs. In the simulated lab, it outperforms alternative approaches to Bayesian optimization in terms of its ability to find optimal configurations, and the number of experiments required to find the optimum. In the real-world lab, the algorithm makes progress toward the optimal setting.
Data availability and implementation: PROTOCOL is available as both a stand-alone Python library, and as part of a R Shiny application at https://github.com/clangmead/PROTOCOL. Data are available at the same repository.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2021. Published by Oxford University Press.
Figures




References
-
- Agrawal S., Goyal N. (2012) Analysis of thompson sampling for the multi-armed bandit problem. In Proc. 25th Annual Conference on Learning Theory, Volume 23 of Proceedings of Machine Learning Research, pp. 39.1–39.26, Edinburgh, Scotland, pp. 25–27.
-
- Bergstra J.S. et al. (2011) Algorithms for hyper-parameter optimization. In: Shawe-Taylor J. et al. (eds.) Advances in Neural Information Processing Systems. Vol. 24. Curran Associates, Inc., Granada, Spain, pp. 2546–2554.
-
- Bubeck S., Cesa-Bianchi N. (2012) Regret analysis of stochastic and nonstochastic multi-armed bandit problems. Found. Trends Mach. Learn., 5, 1–122.
-
- Chang W. et al. (2020) Shiny: Web Application Framework for R. R package version 1.5.0 https://cran.r-project.org/web/packages/shiny/index.htm.
-
- Cummings C.S. et al. (2017) Design of stomach acid-stable and mucin-binding enzyme polymer conjugates. Biomacromolecules, 18, 576–586. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

