EIF4A3-induced circ_0084615 contributes to the progression of colorectal cancer via miR-599/ONECUT2 pathway
- PMID: 34253241
- PMCID: PMC8273970
- DOI: 10.1186/s13046-021-02029-y
EIF4A3-induced circ_0084615 contributes to the progression of colorectal cancer via miR-599/ONECUT2 pathway
Abstract
Background: Colorectal cancer (CRC) is a common malignant tumor. Circular RNAs (circRNAs) have been reported to take part in the progression of CRC. However, the functions of circ_0084615 in CRC development are still undefined. In this study, we aimed to explore the functions and underlying mechanisms of circ_0084615 in CRC.
Methods: qRT-PCR, western blot assay and IHC assay were utilized for the levels of circ_0084615, miR-599, ONECUT2 or EIF4A3. 5-ethynyl-2'-deoxyuridine (EdU) assay and colony formation assay were conducted for cell proliferation ability. Wound-healing assay and transwell assay were applied to evaluate cell migration and invasion. Tube formation assay was used to analyze angiogenesis ability. RNA immunoprecipitation (RIP) assay, RNA pull down assay and dual-luciferase reporter assay were used to analyze the relationships of circ_0084615, miR-599, ONECUT2 and EIF4A3. Murine xenograft model assay was employed for the role of circ_0084615 in vivo.
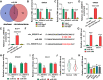
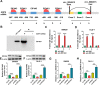
Results: Circ_0084615 was elevated in CRC tissues and was linked to TNM stages, lymph node metastasis, differentiation and overall survival rate. Circ_0084615 knockdown inhibited CRC cell proliferation, migration, invasion and angiogenesis in vitro and hampered tumorigenesis in vivo. Circ_0084615 sponged miR-599 and miR-599 inhibition reversed circ_0084615 knockdown-mediated effects on CRC cell growth, motility and angiogenesis. ONECUT2 was identified as the target gene of miR-599. ONECUT2 overexpression recovered the effects of miR-599 on CRC malignant behaviors. Additionally, EIF4A3 induced circ_0084615 expression.
Conclusions: EIF4A3-induced circ_0084615 played an oncogenic role in CRC development via miR-599/ONECUT2 axis.
Keywords: Colorectal cancer; EIF4A3; ONECUT2; circ_0084615; miR-599.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures








References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

