TSC2 regulates lysosome biogenesis via a non-canonical RAGC and TFEB-dependent mechanism
- PMID: 34253722
- PMCID: PMC8275687
- DOI: 10.1038/s41467-021-24499-6
TSC2 regulates lysosome biogenesis via a non-canonical RAGC and TFEB-dependent mechanism
Abstract
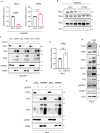
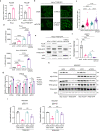
Tuberous Sclerosis Complex (TSC) is caused by TSC1 or TSC2 mutations, resulting in hyperactivation of the mechanistic target of rapamycin complex 1 (mTORC1). Transcription factor EB (TFEB), a master regulator of lysosome biogenesis, is negatively regulated by mTORC1 through a RAG GTPase-dependent phosphorylation. Here we show that lysosomal biogenesis is increased in TSC-associated renal tumors, pulmonary lymphangioleiomyomatosis, kidneys from Tsc2+/- mice, and TSC1/2-deficient cells via a TFEB-dependent mechanism. Interestingly, in TSC1/2-deficient cells, TFEB is hypo-phosphorylated at mTORC1-dependent sites, indicating that mTORC1 is unable to phosphorylate TFEB in the absence of the TSC1/2 complex. Importantly, overexpression of folliculin (FLCN), a GTPase activating protein for RAGC, increases TFEB phosphorylation at the mTORC1 sites in TSC2-deficient cells. Overexpression of constitutively active RAGC is sufficient to relocalize TFEB to the cytoplasm. These findings establish the TSC proteins as critical regulators of lysosomal biogenesis via TFEB and RAGC and identify TFEB as a driver of the proliferation of TSC2-deficient cells.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

