RNA polymerase II speed: a key player in controlling and adapting transcriptome composition
- PMID: 34254686
- PMCID: PMC8327950
- DOI: 10.15252/embj.2020105740
RNA polymerase II speed: a key player in controlling and adapting transcriptome composition
Abstract
RNA polymerase II (RNA Pol II) speed or elongation rate, i.e., the number of nucleotides synthesized per unit of time, is a major determinant of transcriptome composition. It controls co-transcriptional processes such as splicing, polyadenylation, and transcription termination, thus regulating the production of alternative splice variants, circular RNAs, alternatively polyadenylated transcripts, or read-through transcripts. RNA Pol II speed itself is regulated in response to intra- and extra-cellular stimuli and can in turn affect the transcriptome composition in response to these stimuli. Evidence points to a potentially important role of transcriptome composition modification through RNA Pol II speed regulation for adaptation of cells to a changing environment, thus pointing to a function of RNA Pol II speed regulation in cellular physiology. Analyzing RNA Pol II speed dynamics may therefore be central to fully understand the regulation of physiological processes, such as the development of multicellular organisms. Recent findings also raise the possibility that RNA Pol II speed deregulation can be detrimental and participate in disease progression. Here, we review initial and current approaches to measure RNA Pol II speed, as well as providing an overview of the factors controlling speed and the co-transcriptional processes which are affected. Finally, we discuss the role of RNA Pol II speed regulation in cell physiology.
Keywords: RNA polymerase II; co-transcriptional processes; transcription speed.
©2021 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


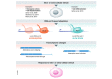
Splicing‐related processes such as constitutive splicing, alternative splicing, and back‐splicing;
The RNA methylome by controlling the deposition of N6‐methyladenosine in RNAs;
Normal 3′ end processing of replication‐dependent core histone mRNAs by controlling the folding of nascent RNA at histone genes;
Termination‐related processes such as alternative polyadenylation (represented by two poly(A) sites) and the distance traveled by RNA Pol II beyond the poly(A) site, i.e., read‐through transcription;
Protein modifications on gene bodies: post‐translational modifications of RNA Pol II CTD such as serine 2 phosphorylation and chromatin modifications such as tri‐methylation of lysine 36 of histone H3.
References
-
- Adkins MW, Tyler JK (2006) Transcriptional activators are dispensable for transcription in the absence of Spt6‐mediated chromatin reassembly of promoter regions. Mol Cell 21: 405–416 - PubMed
-
- Alló M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M , Agirre E, Plass M, Eyras E, Elela SA et al (2009) Control of alternative splicing through siRNA‐mediated transcriptional gene silencing. Nat Struct Mol Biol 16: 717–724 - PubMed
-
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L (2011) Total RNA sequencing reveals nascent transcription and widespread co‐transcriptional splicing in the human brain. Nat Struct Mol Biol 18: 1435–1440 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

