A comparison of epithelial cell content of oral samples estimated using cytology and DNA methylation
- PMID: 34254878
- PMCID: PMC8920143
- DOI: 10.1080/15592294.2021.1950977
A comparison of epithelial cell content of oral samples estimated using cytology and DNA methylation
Abstract
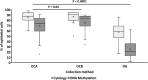
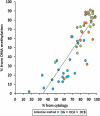
Saliva and buccal samples are popular for epigenome wide association studies (EWAS) due to their ease of collection compared and their ability to sample a different cell lineage compared to blood. As these samples contain a mix of white blood cells and buccal epithelial cells that can vary within a population, this cellular heterogeneity may confound EWAS. This has been addressed by including cellular heterogeneity obtained through cytology at the time of collection or by using cellular deconvolution algorithms built on epigenetic data from specific cell types. However, to our knowledge, the two methods have not yet been compared. Here we show that the two methods are highly correlated in saliva and buccal samples (R = 0.84, P < 0.0001) by comparing data generated from cytological staining and Infinium MethylationEPIC arrays and the EpiDISH deconvolution algorithm from buccal and saliva samples collected from twenty adults. In addition, by using an expanded dataset from both sample types, we confirmed our previous finding that age has strong, non-linear negative correlation with epithelial cell proportion in both sample types. However, children and adults showed a large within-population variation in cellular heterogeneity. Our results validate the use of the EpiDISH algorithm in estimating the effect of cellular heterogeneity in EWAS and showed DNA methylation generally underestimates the epithelial cell content obtained from cytology.
Keywords: Buccal; DNA methylation; EWAS; cell-type heterogeneity; cytology; epithelial cell; saliva.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Teschendorff AE, Zheng SC.. Cell-type deconvolution in epigenome-wide association studies: a review and recommendations. Epigenomics. 2017;9(5):757–768. - PubMed
-
- Teschendorff AE, Relton CL. Statistical and integrative system-level analysis of DNA methylation data. Nat Rev Genet. 2018;19(3):129–147. - PubMed
-
- Zheng SC, Webster AP, Dong D, et al. A novel cell-type deconvolution algorithm reveals substantial contamination by immune cells in saliva, buccal and cervix. Epigenomics. 2018;10(7):925–940. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
