Genome-scale sequencing and analysis of human, wolf, and bison DNA from 25,000-year-old sediment
- PMID: 34256019
- PMCID: PMC8409484
- DOI: 10.1016/j.cub.2021.06.023
Genome-scale sequencing and analysis of human, wolf, and bison DNA from 25,000-year-old sediment
Abstract
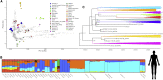
Cave sediments have been shown to preserve ancient DNA but so far have not yielded the genome-scale information of skeletal remains. We retrieved and analyzed human and mammalian nuclear and mitochondrial environmental "shotgun" genomes from a single 25,000-year-old Upper Paleolithic sediment sample from Satsurblia cave, western Georgia:first, a human environmental genome with substantial basal Eurasian ancestry, which was an ancestral component of the majority of post-Ice Age people in the Near East, North Africa, and parts of Europe; second, a wolf environmental genome that is basal to extant Eurasian wolves and dogs and represents a previously unknown, likely extinct, Caucasian lineage; and third, a European bison environmental genome that is basal to present-day populations, suggesting that population structure has been substantially reshaped since the Last Glacial Maximum. Our results provide new insights into the Late Pleistocene genetic histories of these three species and demonstrate that direct shotgun sequencing of sediment DNA, without target enrichment methods, can yield genome-wide data informative of ancestry and phylogenetic relationships.
Keywords: Canis; Caucasus; Upper Paleolithic; bison; enviromental DNA; human; shotgun; soil sequencing.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Comment in
-
Palaeogenetics: Dirt, what is it good for? Everything.Curr Biol. 2021 Aug 23;31(16):R993-R995. doi: 10.1016/j.cub.2021.06.074. Curr Biol. 2021. PMID: 34428419
References
-
- Hagelberg E., Sykes B., Hedges R. Ancient bone DNA amplified. Nature. 1989;342:485. - PubMed
-
- Gilbert M.T.P., Tomsho L.P., Rendulic S., Packard M., Drautz D.I., Sher A., Tikhonov A., Dalén L., Kuznetsova T., Kosintsev P. Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science. 2007;317:1927–1930. - PubMed
-
- Racimo F., Sikora M., Vander Linden M., Schroeder H., Lalueza-Fox C. Beyond broad strokes: sociocultural insights from the study of ancient genomes. Nat. Rev. Genet. 2020;21:355–366. - PubMed
-
- Hofreiter M., Serre D., Poinar H.N., Kuch M., Pääbo S. Ancient DNA. Nat. Rev. Genet. 2001;2:353–359. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

