Neptune: an environment for the delivery of genomic medicine
- PMID: 34257418
- PMCID: PMC8487966
- DOI: 10.1038/s41436-021-01230-w
Neptune: an environment for the delivery of genomic medicine
Abstract
Purpose: Genomic medicine holds great promise for improving health care, but integrating searchable and actionable genetic data into electronic health records (EHRs) remains a challenge. Here we describe Neptune, a system for managing the interaction between a clinical laboratory and an EHR system during the clinical reporting process.
Methods: We developed Neptune and applied it to two clinical sequencing projects that required report customization, variant reanalysis, and EHR integration.
Results: Neptune has been applied for the generation and delivery of over 15,000 clinical genomic reports. This work spans two clinical tests based on targeted gene panels that contain 68 and 153 genes respectively. These projects demanded customizable clinical reports that contained a variety of genetic data types including single-nucleotide variants (SNVs), copy-number variants (CNVs), pharmacogenomics, and polygenic risk scores. Two variant reanalysis activities were also supported, highlighting this important workflow.
Conclusion: Methods are needed for delivering structured genetic data to EHRs. This need extends beyond developing data formats to providing infrastructure that manages the reporting process itself. Neptune was successfully applied on two high-throughput clinical sequencing projects to build and deliver clinical reports to EHR systems. The software is open source and available at https://gitlab.com/bcm-hgsc/neptune .
© 2021. The Author(s), under exclusive licence to the American College of Medical Genetics and Genomics.
Conflict of interest statement
Disclosure: E.V. is a cofounder of Codified Genomics, which provides variant interpretation services. R.G., D.M., D.M., disclose that the Baylor Genetics Laboratory is co-owned by Baylor College of Medicine. All other authors declare no conflicts of interest.
Figures
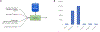
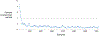
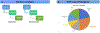
References
-
- Johnson A, Zeng J, Bailey AM, Holla V, Litzenburger B, Lara-Guerra H, Mills GB, Mendelsohn J, Shaw KR, and Meric-Bernstam F (2015). The right drugs at the right time for the right patient: the MD Anderson precision oncology decision support platform. Drug Discovery Today 20, 1433–1438. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- U01 HG008676/HG/NHGRI NIH HHS/United States
- U01 HG008672/HG/NHGRI NIH HHS/United States
- U01 HG008684/HG/NHGRI NIH HHS/United States
- U01 HG008679/HG/NHGRI NIH HHS/United States
- U01 HG008666/HG/NHGRI NIH HHS/United States
- U01 HG008680/HG/NHGRI NIH HHS/United States
- U01 HG008673/HG/NHGRI NIH HHS/United States
- U01 HG008685/HG/NHGRI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- U01 HG008664/HG/NHGRI NIH HHS/United States
- U01 HG008701/HG/NHGRI NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- U01 HG008657/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

