Putative hexameric glycosyltransferase functional unit revealed by the crystal structure of Acinetobacter baumannii MurG
- PMID: 34258006
- PMCID: PMC8256705
- DOI: 10.1107/S2052252521003729
Putative hexameric glycosyltransferase functional unit revealed by the crystal structure of Acinetobacter baumannii MurG
Abstract
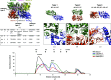
Lipid II, the main component of the bacterial cell wall, is synthesized by the addition of UDP-N-acetylglucosamine to the UDP-N-acetylmuramic acid pentapeptide catalyzed by the glycosyltransferase MurG. Owing to its critical role in cell-wall biosynthesis, MurG is considered to be an attractive target for antibacterial agents. Although the Mur family ligases have been extensively studied, the molecular mechanism of the oligomeric scaffolding assembly of MurG remains unclear. In this study, MurG from Acinetobacter baumannii (abMurG), a human pathogen, was characterized and its hexameric crystal structure was unveiled; this is the first homo-oligomeric structure to be described in the MurG family and the Mur family. Homogeneous protein samples were produced for structural studies using size-exclusion chromatography, the absolute molecular mass was calculated via multi-angle light scattering, and protein-protein interactions were analyzed using the PDBePISA server. abMurG was found to form homo-oligomeric complexes in solution, which might serve as functional units for the scaffolding activity of MurG. Furthermore, analysis of this structure revealed the molecular assembly mechanism of MurG. This structural and biochemical study elucidated the homo-oligomerization mechanism of MurG and suggests a new potential antibiotic target on MurG.
Keywords: Acinetobacter baumannii; MurG; cell-wall peptidoglycan biosynthesis; crystal structure; glycosyltransferases; superbugs.
© Kyoung Ho Jung et al. 2021.
Figures




References
-
- Burki, T. K. (2018). Lancet Respir. Med. 6, 668. - PubMed
LinkOut - more resources
Full Text Sources

