Protocol for improving diffraction quality of leucyl-tRNA synthetase 1 with methylation and post-crystallization soaking and cooling in cryoprotectants
- PMID: 34258600
- PMCID: PMC8260868
- DOI: 10.1016/j.xpro.2021.100642
Protocol for improving diffraction quality of leucyl-tRNA synthetase 1 with methylation and post-crystallization soaking and cooling in cryoprotectants
Abstract
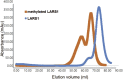
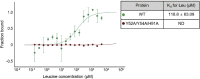
Leucyl-tRNA synthetase 1 (LARS1) synthesizes Leu-tRNALeu for protein synthesis and plays an important role in mTORC1 activation by sensing intracellular leucine concentrations. Here, we describe a protocol for the purification, reductive methylation, binding affinity measurement by microscale thermophoresis, T i value measurement by Tycho, and post-crystallization soaking and cooling in cryoprotectants to improve crystallization of LARS1. Collectively, this allowed us to build the RagD binding domain, which was shown to be a dynamic region of LARS1 refractory to crystallization. For complete details on the use and execution of this protocol, please refer to Kim et al. (2021).
Keywords: Protein Biochemistry; Structural Biology; X-ray Crystallography.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- Adams P.D., Grosse-Kunstleve R.W., Hung L.-W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. - PubMed
-
- Asmari M., Ratih R., Alhazmi H.A., El Deeb S. Thermophoresis for characterizing biomolecular interaction. Methods. 2018;146:107–119. - PubMed
-
- Bartoschik T., Galinec S., Kleusch C., Walkiewicz K., Breitsprecher D., Weigert S., Muller Y.A., You C., Piehler J., Vercruysse T. Near-native, site-specific and purification-free protein labeling for quantitative protein interaction analysis by MicroScale Thermophoresis. Sci. Rep. 2018;8:1–10. - PMC - PubMed
-
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. - PubMed
-
- Han J.M., Jeong S.J., Park M.C., Kim G., Kwon N.H., Kim H.K., Ha S.H., Ryu S.H., Kim S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. - PubMed

