Multidimensional Clinical Surveillance of Pseudomonas aeruginosa Reveals Complex Relationships between Isolate Source, Morphology, and Antimicrobial Resistance
- PMID: 34259555
- PMCID: PMC8386403
- DOI: 10.1128/mSphere.00393-21
Multidimensional Clinical Surveillance of Pseudomonas aeruginosa Reveals Complex Relationships between Isolate Source, Morphology, and Antimicrobial Resistance
Abstract
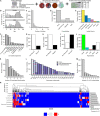
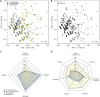
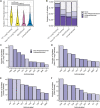
Antimicrobial susceptibility in Pseudomonas aeruginosa is dependent on a complex combination of host and pathogen-specific factors. Through the profiling of 971 clinical P. aeruginosa isolates from 590 patients and collection of paired patient metadata, we show that antimicrobial resistance is associated with not only patient-centric factors (e.g., cystic fibrosis and antipseudomonal prescription history) but also microbe-specific phenotypes (e.g., mucoid colony morphology). Additionally, isolates from different sources (e.g., respiratory tract, urinary tract) displayed rates of antimicrobial resistance that were correlated with source-specific antimicrobial prescription strategies. Furthermore, isolates from the same patient often displayed a high degree of heterogeneity, highlighting a key challenge facing personalized treatment of infectious diseases. Our findings support novel relationships between isolate and patient-level data sets, providing a potential guide for future antimicrobial treatment strategies. IMPORTANCE P. aeruginosa is a leading cause of nosocomial infection and infection in patients with cystic fibrosis. While P. aeruginosa infection and treatment can be complicated by a variety of antimicrobial resistance and virulence mechanisms, pathogen virulence is rarely recorded in a clinical setting. In this study, we discovered novel relationships between antimicrobial resistance, virulence-linked morphologies, and isolate source in a large and variable collection of clinical P. aeruginosa isolates. Our work motivates the clinical surveillance of virulence-linked P. aeruginosa morphologies as well as the tracking of source-specific antimicrobial prescription and resistance patterns.
Keywords: Pseudomonas aeruginosa; antimicrobial resistance; clinical risk factors; infectious disease.
Figures


References
-
- Cystic Fibrosis Foundation. 2019. Cystic Fibrosis Foundation patient registry 2018 annual data report. Cystic Fibrosis Foundation, Bethesda, MD. https://www.cff.org/Research/Researcher-Resources/Patient-Registry/2018-....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

