Gene-Environment Interactions in Developmental Neurotoxicity: a Case Study of Synergy between Chlorpyrifos and CHD8 Knockout in Human BrainSpheres
- PMID: 34259569
- PMCID: PMC8278985
- DOI: 10.1289/EHP8580
Gene-Environment Interactions in Developmental Neurotoxicity: a Case Study of Synergy between Chlorpyrifos and CHD8 Knockout in Human BrainSpheres
Abstract
Background: Autism spectrum disorder (ASD) is a major public health concern caused by complex genetic and environmental components. Mechanisms of gene-environment () interactions and reliable biomarkers associated with ASD are mostly unknown or controversial. Induced pluripotent stem cells (iPSCs) from patients or with clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9 (CRISPR/Cas9)-introduced mutations in candidate ASD genes provide an opportunity to study () interactions.
Objectives: In this study, we aimed to identify a potential synergy between mutation in the high-risk autism gene encoding chromodomain helicase DNA binding protein 8 (CHD8) and environmental exposure to an organophosphate pesticide (chlorpyrifos; CPF) in an iPSC-derived human three-dimensional (3D) brain model.
Methods: This study employed human iPSC-derived 3D brain organoids (BrainSpheres) carrying a heterozygote CRISPR/Cas9-introduced inactivating mutation in CHD8 and exposed to CPF or its oxon-metabolite (CPO). Neural differentiation, viability, oxidative stress, and neurite outgrowth were assessed, and levels of main neurotransmitters and selected metabolites were validated against human data on ASD metabolic derangements.
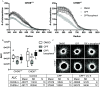
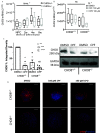
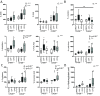
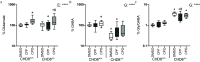
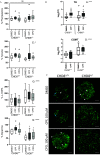
Results: Expression of CHD8 protein was significantly lower in CHD8 heterozygous knockout () BrainSpheres compared with ones. Exposure to CPF/CPO treatment further reduced CHD8 protein levels, showing the potential () interaction synergy. A novel approach for validation of the model was chosen: from the literature, we identified a panel of metabolic biomarkers in patients and assessed them by targeted metabolomics in vitro. A synergistic effect was observed on the cholinergic system, S-adenosylmethionine, S-adenosylhomocysteine, lactic acid, tryptophan, kynurenic acid, and acid levels. Neurite outgrowth was perturbed by CPF/CPO exposure. Heterozygous knockout of CHD8 in BrainSpheres led to an imbalance of excitatory/inhibitory neurotransmitters and lower levels of dopamine.
Discussion: This study pioneered () interaction in iPSC-derived organoids. The experimental strategy enables biomonitoring and environmental risk assessment for ASD. Our findings reflected some metabolic perturbations and disruption of neurotransmitter systems involved in ASD. The increased susceptibility of BrainSpheres to chemical insult establishes a possibly broader role of () interaction in ASD. https://doi.org/10.1289/EHP8580.
Figures


Comment in
-
Autism in Three Dimensions: Using Brain Organoids to Study Potential Gene-Environment Interactions.Environ Health Perspect. 2021 Oct;129(10):104003. doi: 10.1289/EHP10301. Epub 2021 Oct 19. Environ Health Perspect. 2021. PMID: 34665007 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

