The Baltimore Classification of Viruses 50 Years Later: How Does It Stand in the Light of Virus Evolution?
- PMID: 34259570
- PMCID: PMC8483701
- DOI: 10.1128/MMBR.00053-21
The Baltimore Classification of Viruses 50 Years Later: How Does It Stand in the Light of Virus Evolution?
Abstract
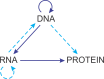
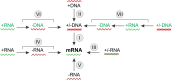
Fifty years ago, David Baltimore published a brief conceptual paper delineating the classification of viruses by the routes of genome expression. The six "Baltimore classes" of viruses, with a subsequently added 7th class, became the conceptual framework for the development of virology during the next five decades. During this time, it became clear that the Baltimore classes, with relatively minor additions, indeed cover the diversity of virus genome expression schemes that also define the replication cycles. Here, we examine the status of the Baltimore classes 50 years after their advent and explore their links with the global ecology and biology of the respective viruses. We discuss an extension of the Baltimore scheme and why many logically admissible expression-replication schemes do not appear to be realized in nature. Recent phylogenomic analyses allow tracing the complex connections between the Baltimore classes and the monophyletic realms of viruses. The five classes of RNA viruses and reverse-transcribing viruses share an origin, whereas both the single-stranded DNA viruses and double-stranded DNA (dsDNA) viruses evolved on multiple independent occasions. Most of the Baltimore classes of viruses probably emerged during the earliest era of life evolution, at the stage of the primordial pool of diverse replicators, and before the advent of modern-like cells with large dsDNA genomes. The Baltimore classes remain an integral part of the conceptual foundation of biology, providing the essential structure for the logical space of information transfer processes, which is nontrivially connected with the routes of evolution of viruses and other replicators.
Keywords: virus classification; virus evolution; virus realms; virus taxonomy.
Figures



References
-
- Baltimore D. 1974. The strategy of RNA viruses. Harvey Lect 70 Series: 57–74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

