Structure and Zeatin Binding of the Peach Allergen Pru p 1
- PMID: 34260238
- PMCID: PMC8323099
- DOI: 10.1021/acs.jafc.1c01876
Structure and Zeatin Binding of the Peach Allergen Pru p 1
Abstract
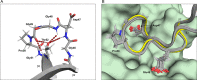
Peach (Prunus persica) is among the fruits most frequently reported to cause food allergies. Allergic reactions commonly result from previous sensitization to the birch pollen allergen Bet v 1, followed by immunological cross-reactivity of IgE antibodies to structurally related proteins in peach. In this study, we present the three-dimensional NMR solution structure of the cross-reactive peach allergen Pru p 1 (isoform Pru p 1.0101). This 17.5 kDa protein adopts the canonical Bet v 1 fold, composed of a seven-stranded β-sheet and three α-helices enclosing an internal cavity. In Pru p 1, the inner surface of the cavity contains an array of hydroxyl-bearing amino acids surrounded by a hydrophobic patch, constituting a docking site for amphiphilic molecules. NMR-guided docking of the cytokinin molecule zeatin to the internal cavity of Pru p 1 provides a structure-based rationale for the effect that zeatin binding has on the protein's RNase activity.
Keywords: NMR structure; allergen; cytokinin; ribonuclease.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Burney P. G. J.; Potts J.; Kummeling I.; Mills E. N. C.; Clausen M.; Dubakiene R.; Barreales L.; Fernandez-Perez C.; Fernandez-Rivas M.; Le T.-M.; Knulst A. C.; Kowalski M. L.; Lidholm J.; Ballmer-Weber B. K.; Braun-Fahlander C.; Mustakov T.; Kralimarkova T.; Popov T.; Sakellariou A.; Papadopoulos N. G.; Versteeg S. A.; Zuidmeer L.; Akkerdaas J. H.; Hoffmann-Sommergruber K.; van Ree R. The prevalence and distribution of food sensitization in European adults. Allergy 2014, 69, 365–371. 10.1111/all.12341. - DOI - PubMed
-
- Geroldinger-Simic M.; Zelniker T.; Aberer W.; Ebner C.; Egger C.; Greiderer A.; Prem N.; Lidholm J.; Ballmer-Weber B. K.; Vieths S.; Bohle B. Birch pollen-related food allergy: clinical aspects and the role of allergen-specific IgE and IgG4 antibodies. J. Allergy Clin. Immunol. 2011, 127, 616–622. 10.1016/j.jaci.2010.10.027. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

