Nutritional regulation of oligodendrocyte differentiation regulates perineuronal net remodeling in the median eminence
- PMID: 34260928
- PMCID: PMC8293628
- DOI: 10.1016/j.celrep.2021.109362
Nutritional regulation of oligodendrocyte differentiation regulates perineuronal net remodeling in the median eminence
Abstract
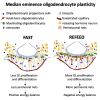
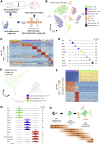
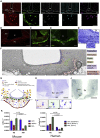
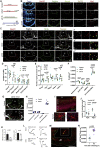
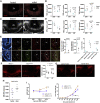
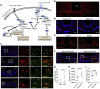
The mediobasal hypothalamus (MBH; arcuate nucleus of the hypothalamus [ARH] and median eminence [ME]) is a key nutrient sensing site for the production of the complex homeostatic feedback responses required for the maintenance of energy balance. Here, we show that refeeding after an overnight fast rapidly triggers proliferation and differentiation of oligodendrocyte progenitors, leading to the production of new oligodendrocytes in the ME specifically. During this nutritional paradigm, ME perineuronal nets (PNNs), emerging regulators of ARH metabolic functions, are rapidly remodeled, and this process requires myelin regulatory factor (Myrf) in oligodendrocyte progenitors. In genetically obese ob/ob mice, nutritional regulations of ME oligodendrocyte differentiation and PNN remodeling are blunted, and enzymatic digestion of local PNN increases food intake and weight gain. We conclude that MBH PNNs are required for the maintenance of energy balance in lean mice and are remodeled in the adult ME by the nutritional control of oligodendrocyte differentiation.
Keywords: energy balance; glucose homeostasis; hypothalamus; median eminence; nutrition; obesity; oligodendrocyte; perineuronal nets; plasticity.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Alonge K.M., Mirzadeh Z., Scarlett J.M., Logsdon A.F., Brown J.M., Cabrales E., Chan C.K., Kaiyala K.J., Bentsen M.A., Banks W.A. Hypothalamic perineuronal net assembly is required for sustained diabetes remission induced by fibroblast growth factor 1 in rats. Nat. Metab. 2020;2:1025–1033. - PMC - PubMed
-
- Bayraktar O.A., Bartels T., Polioudakis D., Holmqvist S., Haim L.B., Young A.M.H., Prakash K., Brown A., Paredes M.F., Kawaguchi R. Single-cell in situ transcriptomic map of astrocyte cortical layer diversity. bioRxiv. 2018 doi: 10.1101/432104. - DOI
-
- Bibollet-Bahena O., Almazan G. IGF-1-stimulated protein synthesis in oligodendrocyte progenitors requires PI3K/mTOR/Akt and MEK/ERK pathways. J. Neurochem. 2009;109:1440–1451. - PubMed
-
- Bin J.M., Harris S.N., Kennedy T.E. The oligodendrocyte-specific antibody ‘CC1’ binds Quaking 7. J. Neurochem. 2016;139:181–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 108926/B/15/Z/WT_/Wellcome Trust/United Kingdom
- 204488/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MC_UU_00014/5/MRC_/Medical Research Council/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/S026193/1/MRC_/Medical Research Council/United Kingdom
- MR/S011552/1/MRC_/Medical Research Council/United Kingdom
- MRC_MC_UU_00014/5/MRC_/Medical Research Council/United Kingdom
- 100574/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- MC_UU_00014/1/MRC_/Medical Research Council/United Kingdom
- 208363/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- MC_PC_17230/MRC_/Medical Research Council/United Kingdom
- T32 HL007028/HL/NHLBI NIH HHS/United States
- MC_UU_12012/1/MRC_/Medical Research Council/United Kingdom
- R01 DK083042/DK/NIDDK NIH HHS/United States
- MC_UU_12012/5/MRC_/Medical Research Council/United Kingdom
- MRC_MC_UU_12012/5/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

