Adaptive immunity induces mutualism between commensal eukaryotes
- PMID: 34262174
- PMCID: PMC8904204
- DOI: 10.1038/s41586-021-03722-w
Adaptive immunity induces mutualism between commensal eukaryotes
Abstract
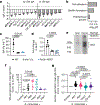
Pathogenic fungi reside in the intestinal microbiota but rarely cause disease. Little is known about the interactions between fungi and the immune system that promote commensalism. Here we investigate the role of adaptive immunity in promoting mutual interactions between fungi and host. We find that potentially pathogenic Candida species induce and are targeted by intestinal immunoglobulin A (IgA) responses. Focused studies on Candida albicans reveal that the pathogenic hyphal morphotype, which is specialized for adhesion and invasion, is preferentially targeted and suppressed by intestinal IgA responses. IgA from mice and humans directly targets hyphal-enriched cell-surface adhesins. Although typically required for pathogenesis, C. albicans hyphae are less fit for gut colonization1,2 and we show that immune selection against hyphae improves the competitive fitness of C. albicans. C. albicans exacerbates intestinal colitis3 and we demonstrate that hyphae and an IgA-targeted adhesin exacerbate intestinal damage. Finally, using a clinically relevant vaccine to induce an adhesin-specific immune response protects mice from C. albicans-associated damage during colitis. Together, our findings show that adaptive immunity suppresses harmful fungal effectors, with benefits to both C. albicans and its host. Thus, IgA uniquely uncouples colonization from pathogenesis in commensal fungi to promote homeostasis.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures






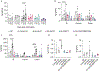
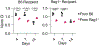



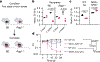

Comment in
-
IgA selects for fit, peace-loving fungi.Nat Rev Immunol. 2021 Sep;21(9):546-547. doi: 10.1038/s41577-021-00594-z. Nat Rev Immunol. 2021. PMID: 34302140 No abstract available.
References
-
- Tso GHW et al. Experimental evolution of a fungal pathogen into a gut symbiont. Science 362, 589–595 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI141202/AI/NIAID NIH HHS/United States
- R01 AG047956/AG/NIA NIH HHS/United States
- S10 OD018210/OD/NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- DP2 GM111099/GM/NIGMS NIH HHS/United States
- R01 AI046223/AI/NIAID NIH HHS/United States
- T32 AI055434/AI/NIAID NIH HHS/United States
- S10 OD026959/OD/NIH HHS/United States
- 5P30 CA042014-24/NH/NIH HHS/United States
- R01 DK124336/DK/NIDDK NIH HHS/United States
- R01 AT011423/AT/NCCIH NIH HHS/United States
- R00 HL102228-05/NH/NIH HHS/United States
- P30 CA042014/CA/NCI NIH HHS/United States
- DP2 GM111099-01/NH/NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AI063503/AI/NIAID NIH HHS/United States
- R00 HL102228/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

