Analysis of Age-Related Circular RNA Expression Profiles in Mesenchymal Stem Cells of Rat Bone Marrow
- PMID: 34262589
- PMCID: PMC8273880
- DOI: 10.3389/fgene.2021.600632
Analysis of Age-Related Circular RNA Expression Profiles in Mesenchymal Stem Cells of Rat Bone Marrow
Abstract
As multicellular organisms age, they undergo a reduction in tissue and organ function. Researchers have put forward a theory that stem cell aging is the main factor responsible for decreased tissue and organ function. The adult stem cells guarantee the maintenance and repair of adult tissues and organs. Among adult stem cells, mesenchymal stem cells (MSCs) are emerging as hopeful candidates for cell-based therapy of numerous diseases. In recent years, high-throughput sequencing technologies have evolved to identify circular RNAs (circRNAs) associated with an increasing number of diseases, such as cancer and age-related diseases. It has been reported that circRNAs can compete with microRNAs (miRNAs) to affect the stability or translation of target RNAs and further regulate gene expression at the transcriptional level. However, the role of circRNAs expressed in MSCs in aging mechanisms has not yet been deciphered. The aim of this study was to explore and analyze the expression profiles of age-related circRNAs in MSCs. In this study, bone marrow MSCs were extracted from aged and young rats and analyzed using high-throughput sequencing and bioinformatics. The reliability of high-throughput RNA sequencing was verified by quantitative real-time polymerase chain reaction. The most important circRNA functions and pathways were further selected by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomics (KEGG) analysis. Age-related circRNAs were found in the circrNA-miRNA-mRNA interaction network. The results of high-throughput sequencing showed that 4,229 circRNAs were involved in age-related senescence of MSCs. Compared with the young group, there were 29 differentially expressed circRNAs in the aged group, of which four were upregulated and 25 were downregulated. GO analysis covered three domains: biological process (BP), cellular component (CC), and molecular function (MF). The terms assigned to the BP domain were cellular metabolic processes and cellular macromolecule metabolic processes. The identified CC terms were intracellular and intracellular part, and the identified MF terms were binding and protein binding. The top five KEGG pathways were mitophagy-animal-Rattus norvegicus, prostate cancer-Rattus norvegicus, pathways in cancer-Rattus norvegicus, lysosome-Rattus norvegicus, and autophagy-animal-Rattus norvegicus. Altogether, circRNAs may play a major role in age-related MSC senescence. This study provides new mechanistic insights into MSC senescence, possibly leading to novel therapeutic strategies for age-related diseases.
Keywords: MSCs; bioinformatics; circular RNA; high-throughput sequencing; senescence.
Copyright © 2021 Sun, Sun, Yu, Gao, Wang, Zhang, Shi and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


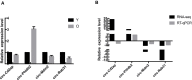




References
-
- Banfi A., Muraglia A., Dozin B., Mastrogiacomo M., Cancedda R., Quarto R. (2000). Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: implications for their use in cell therapy. Exp. Hematol. 28 707–715. 10.1016/S0301-472x(00)00160-0 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

