Identification of a novel lineage bat SARS-related coronaviruses that use bat ACE2 receptor
- PMID: 34263709
- PMCID: PMC8344244
- DOI: 10.1080/22221751.2021.1956373
Identification of a novel lineage bat SARS-related coronaviruses that use bat ACE2 receptor
Abstract
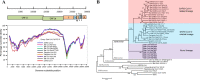
Severe respiratory disease coronavirus-2 (SARS-CoV-2) has been the most devastating disease COVID-19 in the century. One of the unsolved scientific questions of SARS-CoV-2 is the animal origin of this virus. Bats and pangolins are recognized as the most probable reservoir hosts that harbour highly similar SARS-CoV-2 related viruses (SARSr-CoV-2). This study identified a novel lineage of SARSr-CoVs, including RaTG15 and seven other viruses, from bats at the same location where we found RaTG13 in 2015. Although RaTG15 and the related viruses share 97.2% amino acid sequence identities with SARS-CoV-2 in the conserved ORF1b region, it only shows less than 77.6% nucleotide identity to all known SARSr-CoVs at the genome level, thus forming a distinct lineage in the Sarbecovirus phylogenetic tree. We found that the RaTG15 receptor-binding domain (RBD) can bind to ACE2 from Rhinolophus affinis, Malayan pangolin, and use it as an entry receptor, except for ACE2 from humans. However, it contains a short deletion and has different key residues responsible for ACE2 binding. In addition, we showed that none of the known viruses in bat SARSr-CoV-2 lineage discovered uses human ACE2 as efficiently as the pangolin-derived SARSr-CoV-2 or some viruses in the SARSr-CoV-1 lineage. Therefore, further systematic and longitudinal studies in bats are needed to prevent future spillover events caused by SARSr-CoVs or to understand the origin of SARS-CoV-2 better.
Keywords: ACE2; SARS-related coronavirus; bat; novel lineage; reservoir host.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Zhou P, Shi ZL.. SARS-CoV-2 spillover events. Science. 2021;371(6525):120–122. - PubMed
-
- Xiao K, Zhai J, Feng Y, et al. . Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583(7815):286–289. - PubMed
-
- Lam TT-Y, Jia N, Zhang Y-W, et al. . Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583(7815):282–285. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
