Endothelium-protective, histone-neutralizing properties of the polyanionic agent defibrotide
- PMID: 34264868
- PMCID: PMC8492316
- DOI: 10.1172/jci.insight.149149
Endothelium-protective, histone-neutralizing properties of the polyanionic agent defibrotide
Abstract
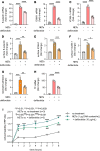
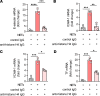
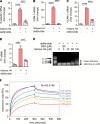
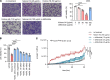
Neutrophil-mediated activation and injury of the endothelium play roles in the pathogenesis of diverse disease states ranging from autoimmunity to cancer to COVID-19. Neutralization of cationic proteins (such as neutrophil extracellular trap-derived [NET-derived] histones) with polyanionic compounds has been suggested as a potential strategy for protecting the endothelium from such insults. Here, we report that the US Food and Drug Administration-approved polyanionic agent defibrotide (a pleiotropic mixture of oligonucleotides) directly engages histones and thereby blocks their pathological effects on endothelium. In vitro, defibrotide counteracted endothelial cell activation and pyroptosis-mediated cell death, whether triggered by purified NETs or recombinant histone H4. In vivo, defibrotide stabilized the endothelium and protected against histone-accelerated inferior vena cava thrombosis in mice. Mechanistically, defibrotide demonstrated direct and tight binding to histone H4 as detected by both electrophoretic mobility shift assay and surface plasmon resonance. Taken together, these data provide insights into the potential role of polyanionic compounds in protecting the endothelium from thromboinflammation with potential implications for myriad NET- and histone-accelerated disease states.
Keywords: Endothelial cells; Inflammation; Neutrophils; Thrombosis; Vascular Biology.
Conflict of interest statement
Figures



Update of
-
Endothelium-protective, histone-neutralizing properties of the polyanionic agent defibrotide.medRxiv [Preprint]. 2021 Jul 15:2021.02.21.21252160. doi: 10.1101/2021.02.21.21252160. medRxiv. 2021. Update in: JCI Insight. 2021 Sep 8;6(17):149149. doi: 10.1172/jci.insight.149149. PMID: 33655266 Free PMC article. Updated. Preprint.
Comment in
-
Defibrotide inhibits NET-mediated thrombosis in APS models.Nat Rev Rheumatol. 2022 Feb;18(2):63. doi: 10.1038/s41584-021-00742-8. Nat Rev Rheumatol. 2022. PMID: 34949770 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

