Renewed interests in the discovery of bioactive actinomycete metabolites driven by emerging technologies
- PMID: 34265147
- PMCID: PMC8714619
- DOI: 10.1111/jam.15225
Renewed interests in the discovery of bioactive actinomycete metabolites driven by emerging technologies
Abstract
Actinomycetes are prolific sources of bioactive molecules. Traditional workflows including bacterial isolation, fermentation, metabolite identification and structure elucidation have resulted in high rates of natural product rediscovery in recent years. Recent advancements in multi-omics techniques have uncovered cryptic gene clusters within the genomes of actinomycetes, potentially introducing vast resources for the investigation of bioactive molecules. While developments in culture techniques have allowed for the fermentation of difficult-to-culture actinomycetes, high-throughput metabolite screening has offered plenary tools to accelerate hits discovery. A variety of new bioactive molecules have been isolated from actinomycetes of unique environmental origins, such as endophytic and symbiotic actinomycetes. Synthetic biology and genome mining have also emerged as new frontiers for the discovery of bioactive molecules. This review covers the highlights of recent developments in actinomycete-derived natural product drug discovery.
Keywords: Streptomyces; actinomycetes; bioactive metabolites; co-culture; endophytes; multi-omics.
© 2021 The Society for Applied Microbiology.
Conflict of interest statement
No conflict of interest declared.
Conflict of Interest
The authors declare that there is no conflict of interest.
Figures



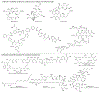
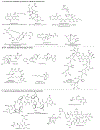

References
-
- Adnani N, Chevrette MG, Adibhatla SN, Zhang F, Yu Q, Braun DR, Nelson J, Simpkins SW, Mcdonald BR, Myers CL, Piotrowski JS, Thompson CJ, Currie CR, Li L, Rajski SR and Bugni TS (2017). Coculture of Marine Invertebrate-Associated Bacteria and Interdisciplinary Technologies Enable Biosynthesis and Discovery of a New Antibiotic, Keyicin. ACS Chem Biol 12, 3093–3102. - PMC - PubMed
-
- Aroonsri A, Kitani S, Choi SU and Nihira T (2008). Isolation and characterization of bamA genes, homologues of the gamma-butyrolactone autoregulator-receptor gene in Amycolatopsis mediterranei, a rifamycin producer. Biotechnol Lett 30, 2019–2024. - PubMed
-
- Arsic B, Barber J, Čikoš A, Mladenovic M, Stankovic N and Novak P (2018). 16-membered macrolide antibiotics: a review. Int J Antimicrob Agents 51, 283–298. - PubMed

