Global analysis of protein lysine 2-hydroxyisobutyrylation (Khib) profiles in Chinese herb rhubarb (Dahuang)
- PMID: 34266380
- PMCID: PMC8283887
- DOI: 10.1186/s12864-021-07847-0
Global analysis of protein lysine 2-hydroxyisobutyrylation (Khib) profiles in Chinese herb rhubarb (Dahuang)
Abstract
Background: Lysine 2-hydroxyisobutyrylation (Khib) is a newly discovered protein posttranslational modification (PTM) and is involved in the broad-spectrum regulation of cellular processes that are found in both prokaryotic and eukaryotic cells, including in plants. The Chinese herb rhubarb (Dahuang) is one of the most widely used traditional Chinese medicines in clinical applications. To better understand the physiological activities and mechanism of treating diseases with the herb, it is necessary to conduct intensive research on rhubarb. However, Khib modification has not been reported thus far in rhubarb.
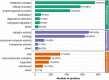
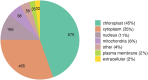
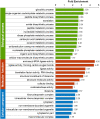
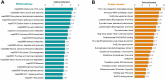
Results: In this study, we performed the first global analysis of Khib-modified proteins in rhubarb by using sensitive affinity enrichment combined with high-accuracy HPLC-MS/MS tandem spectrometry. A total of 4333 overlapping Khib modification peptides matched on 1525 Khib-containing proteins were identified in three independent tests. Bioinformatics analysis showed that these Khib-containing proteins are involved in a wide range of cellular processes, particularly in protein biosynthesis and central carbon metabolism and are distributed mainly in chloroplasts, cytoplasm, nucleus and mitochondria. In addition, the amino acid sequence motif analysis showed that a negatively charged side chain residue (E), a positively charged residue (K), and an uncharged residue with the smallest side chain (G) were strongly preferred around the Khib site, and a total of 13 Khib modification motifs were identified. These identified motifs can be classified into three motif patterns, and some motif patterns are unique to rhubarb and have not been identified in other plants to date.
Conclusions: A total of 4333 Khib-modified peptides on 1525 proteins were identified. The Khib-modified proteins are mainly distributed in the chloroplast, cytoplasm, nucleus and mitochondria, and involved in a wide range of cellular processes. Moreover, three types of amino acid sequence motif patterns, including EKhib/KhibE, GKhib and k.kkk….Khib….kkkkk, were extracted from a total of 13 Khib-modified peptides. This study provides comprehensive Khib-proteome resource of rhubarb. The findings from the study contribute to a better understanding of the physiological roles of Khib modification, and the Khib proteome data will facilitate further investigations of the roles and mechanisms of Khib modification in rhubarb.
Keywords: Chinese herb rhubarb (Dahuang); Lysine 2-hydroxyisobutyrylation (Khib); Motif; Posttranslational modification (PTM).
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

