Multi-institutional TSA-amplified Multiplexed Immunofluorescence Reproducibility Evaluation (MITRE) Study
- PMID: 34266881
- PMCID: PMC8286792
- DOI: 10.1136/jitc-2020-002197
Multi-institutional TSA-amplified Multiplexed Immunofluorescence Reproducibility Evaluation (MITRE) Study
Abstract
Background: Emerging data suggest predictive biomarkers based on the spatial arrangement of cells or coexpression patterns in tissue sections will play an important role in precision immuno-oncology. Multiplexed immunofluorescence (mIF) is ideally suited to such assessments. Standardization and validation of an end-to-end workflow that supports multisite trials and clinical laboratory processes are vital. Six institutions collaborated to: (1) optimize an automated six-plex assay focused on the PD-1/PD-L1 axis, (2) assess intersite and intrasite reproducibility of staining using a locked down image analysis algorithm to measure tumor cell and immune cell (IC) subset densities, %PD-L1 expression on tumor cells (TCs) and ICs, and PD-1/PD-L1 proximity assessments.
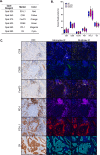
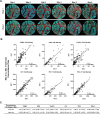
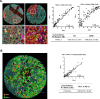
Methods: A six-plex mIF panel (PD-L1, PD-1, CD8, CD68, FOXP3, and CK) was rigorously optimized as determined by quantitative equivalence to immunohistochemistry (IHC) chromogenic assays. Serial sections from tonsil and breast carcinoma and non-small cell lung cancer (NSCLC) tissue microarrays (TMAs), TSA-Opal fluorescent detection reagents, and antibodies were distributed to the six sites equipped with a Leica Bond Rx autostainer and a Vectra Polaris multispectral imaging platform. Tissue sections were stained and imaged at each site and delivered to a single site for analysis. Intersite and intrasite reproducibility were assessed by linear fits to plots of cell densities, including %PDL1 expression by TCs and ICs in the breast and NSCLC TMAs.
Results: Comparison of the percent positive cells for each marker between mIF and IHC revealed that enhanced amplification in the mIF assay was required to detect low-level expression of PD-1, PD-L1, FoxP3 and CD68. Following optimization, an average equivalence of 90% was achieved between mIF and IHC across all six assay markers. Intersite and intrasite cell density assessments showed an average concordance of R2=0.75 (slope=0.92) and R2=0.88 (slope=0.93) for breast carcinoma, respectively, and an average concordance of R2=0.72 (slope=0.86) and R2=0.81 (slope=0.68) for NSCLC. Intersite concordance for %PD-L1+ICs had an average R2 value of 0.88 and slope of 0.92. Assessments of PD-1/PD-L1 proximity also showed strong concordance (R2=0.82; slope=0.75).
Conclusions: Assay optimization yielded highly sensitive, reproducible mIF characterization of the PD-1/PD-L1 axis across multiple sites. High concordance was observed across sites for measures of density of specific IC subsets, measures of coexpression and proximity with single-cell resolution.
Keywords: biomarkers; breast neoplasms; immunohistochemistry; lung neoplasms; programmed cell death 1 receptor; tumor.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: CB-M, SMJ, and BAF: research support from Bristol Myers Squibb II-ON program, and equipment and supply support from Akoya Biosciences; JT: research support from Bristol Myers Squibb; DLR declares that in the last 2 years, he has served as a consultant to AstraZeneca, Amgen, BMS, Cell Signaling Technology, Cepheid, Daiichi Sankyo, Danaher, GSK, Konica/Minolta, Merck, NanoString, Novartis, PAIGE.AI, PerkinElmer/Akoya Biosciences, Ultivue, and Ventana Medical Systems; BAF declares consulting for Ultivue and Neogenomics and research support from Macrogenics, Bristol Myers Squibb, Incyte, OncoSec Medical, and Merck; KR, CW, JM, CC, BR, DL, and CH: all are employees of Akoya Biosciences. No potential conflicts of interest were disclosed by the other authors.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
