Tree-aggregated predictive modeling of microbiome data
- PMID: 34267244
- PMCID: PMC8282688
- DOI: 10.1038/s41598-021-93645-3
Tree-aggregated predictive modeling of microbiome data
Abstract
Modern high-throughput sequencing technologies provide low-cost microbiome survey data across all habitats of life at unprecedented scale. At the most granular level, the primary data consist of sparse counts of amplicon sequence variants or operational taxonomic units that are associated with taxonomic and phylogenetic group information. In this contribution, we leverage the hierarchical structure of amplicon data and propose a data-driven and scalable tree-guided aggregation framework to associate microbial subcompositions with response variables of interest. The excess number of zero or low count measurements at the read level forces traditional microbiome data analysis workflows to remove rare sequencing variants or group them by a fixed taxonomic rank, such as genus or phylum, or by phylogenetic similarity. By contrast, our framework, which we call trac (tree-aggregation of compositional data), learns data-adaptive taxon aggregation levels for predictive modeling, greatly reducing the need for user-defined aggregation in preprocessing while simultaneously integrating seamlessly into the compositional data analysis framework. We illustrate the versatility of our framework in the context of large-scale regression problems in human gut, soil, and marine microbial ecosystems. We posit that the inferred aggregation levels provide highly interpretable taxon groupings that can help microbiome researchers gain insights into the structure and functioning of the underlying ecosystem of interest.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

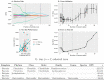
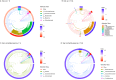

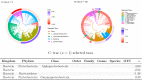
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

