DeepEMhancer: a deep learning solution for cryo-EM volume post-processing
- PMID: 34267316
- PMCID: PMC8282847
- DOI: 10.1038/s42003-021-02399-1
DeepEMhancer: a deep learning solution for cryo-EM volume post-processing
Abstract
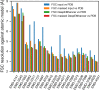
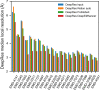
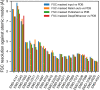
Cryo-EM maps are valuable sources of information for protein structure modeling. However, due to the loss of contrast at high frequencies, they generally need to be post-processed to improve their interpretability. Most popular approaches, based on global B-factor correction, suffer from limitations. For instance, they ignore the heterogeneity in the map local quality that reconstructions tend to exhibit. Aiming to overcome these problems, we present DeepEMhancer, a deep learning approach designed to perform automatic post-processing of cryo-EM maps. Trained on a dataset of pairs of experimental maps and maps sharpened using their respective atomic models, DeepEMhancer has learned how to post-process experimental maps performing masking-like and sharpening-like operations in a single step. DeepEMhancer was evaluated on a testing set of 20 different experimental maps, showing its ability to reduce noise levels and obtain more detailed versions of the experimental maps. Additionally, we illustrated the benefits of DeepEMhancer on the structure of the SARS-CoV-2 RNA polymerase.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

