MASiVar: Multisite, multiscanner, and multisubject acquisitions for studying variability in diffusion weighted MRI
- PMID: 34270123
- PMCID: PMC9087815
- DOI: 10.1002/mrm.28926
MASiVar: Multisite, multiscanner, and multisubject acquisitions for studying variability in diffusion weighted MRI
Abstract
Purpose: Diffusion-weighted imaging allows investigators to identify structural, microstructural, and connectivity-based differences between subjects, but variability due to session and scanner biases is a challenge.
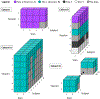
Methods: To investigate DWI variability, we present MASiVar, a multisite data set consisting of 319 diffusion scans acquired at 3 T from b = 1000 to 3000 s/mm2 across 14 healthy adults, 83 healthy children (5 to 8 years), three sites, and four scanners as a publicly available, preprocessed, and de-identified data set. With the adult data, we demonstrate the capacity of MASiVar to simultaneously quantify the intrasession, intersession, interscanner, and intersubject variability of four common DWI processing approaches: (1) a tensor signal representation, (2) a multi-compartment neurite orientation dispersion and density model, (3) white-matter bundle segmentation, and (4) structural connectomics. Respectively, we evaluate region-wise fractional anisotropy, mean diffusivity, and principal eigenvector; region-wise CSF volume fraction, intracellular volume fraction, and orientation dispersion index; bundle-wise shape, volume, fractional anisotropy, and length; and whole connectome correlation and maximized modularity, global efficiency, and characteristic path length.
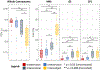
Results: We plot the variability in these measures at each level and find that it consistently increases with intrasession to intersession to interscanner to intersubject effects across all processing approaches and that sometimes interscanner variability can approach intersubject variability.
Conclusions: This study demonstrates the potential of MASiVar to more globally investigate DWI variability across multiple levels and processing approaches simultaneously and suggests harmonization between scanners for multisite analyses should be considered before inference of group differences on subjects.
Keywords: DTI; NODDI; bundle segmentation; connectome; reproducibility; variability.
© 2021 International Society for Magnetic Resonance in Medicine.
Figures






References
-
- Assaf Y, Pasternak O. Diffusion tensor imaging (DTI)-based white matter mapping in brain research: a review. J Mol Neurosci. 2008;34:51–61. - PubMed
-
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012;61:1000–1016. - PubMed
-
- Stanisz GJ, Szafer A, Wright GA, Henkelman RM. An analytical model of restricted diffusion in bovine optic nerve. Magn Reson Med. 1997;37:103–111. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

