Transcriptomic Profiling of Canine Atrial Fibrillation Models After One Week of Sustained Arrhythmia
- PMID: 34270327
- PMCID: PMC8376273
- DOI: 10.1161/CIRCEP.121.009887
Transcriptomic Profiling of Canine Atrial Fibrillation Models After One Week of Sustained Arrhythmia
Erratum in
-
Correction to: Transcriptomic Profiling of Canine Atrial Fibrillation Models After One Week of Sustained Arrhythmia.Circ Arrhythm Electrophysiol. 2021 Sep;14(9):e000079. doi: 10.1161/HAE.0000000000000079. Epub 2021 Sep 21. Circ Arrhythm Electrophysiol. 2021. PMID: 34546789 Free PMC article. No abstract available.
Abstract
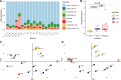
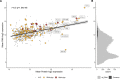
[Figure: see text].
Keywords: atrial fibrillation; atrial remodeling; glutamate; microRNAs; transcriptomics.
Figures



References
-
- Andrade J, Khairy P, Dobrev D, Nattel S. The clinical profile and pathophysiology of atrial fibrillation: relationships among clinical features, epidemiology, and mechanisms. Circ Res. 2014;114:1453–1468. doi: 10.1161/CIRCRESAHA.114.303211 - PubMed
-
- Wijffels MC, Kirchhof CJ, Dorland R, Allessie MA. Atrial fibrillation begets atrial fibrillation. A study in awake chronically instrumented goats. Circulation. 1995;92:1954–1968. doi: 10.1161/01.cir.92.7.1954 - PubMed
-
- Nattel S, Harada M. Atrial remodeling and atrial fibrillation: recent advances and translational perspectives. J Am Coll Cardiol. 2014;63:2335–2345. doi: 10.1016/j.jacc.2014.02.555 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

