Large-scale whole-genome resequencing unravels the domestication history of Cannabis sativa
- PMID: 34272249
- PMCID: PMC8284894
- DOI: 10.1126/sciadv.abg2286
Large-scale whole-genome resequencing unravels the domestication history of Cannabis sativa
Abstract
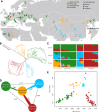
Cannabis sativa has long been an important source of fiber extracted from hemp and both medicinal and recreational drugs based on cannabinoid compounds. Here, we investigated its poorly known domestication history using whole-genome resequencing of 110 accessions from worldwide origins. We show that C. sativa was first domesticated in early Neolithic times in East Asia and that all current hemp and drug cultivars diverged from an ancestral gene pool currently represented by feral plants and landraces in China. We identified candidate genes associated with traits differentiating hemp and drug cultivars, including branching pattern and cellulose/lignin biosynthesis. We also found evidence for loss of function of genes involved in the synthesis of the two major biochemically competing cannabinoids during selection for increased fiber production or psychoactive properties. Our results provide a unique global view of the domestication of C. sativa and offer valuable genomic resources for ongoing functional and molecular breeding research.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures


References
-
- Small E., Evolution and classification of Cannabis sativa (marijuana, hemp) in relation to human utilization. Bot. Rev. 81, 189–294 (2015).
-
- R. C. Clarke, M. D. Merlin, Cannabis—Evolution and Ethnobotany (University of California Press, 2013).
LinkOut - more resources
Full Text Sources
Other Literature Sources

