Comprehensive functional evaluation of variants of fibroblast growth factor receptor genes in cancer
- PMID: 34272467
- PMCID: PMC8285406
- DOI: 10.1038/s41698-021-00204-0
Comprehensive functional evaluation of variants of fibroblast growth factor receptor genes in cancer
Abstract
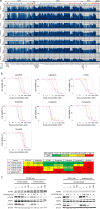
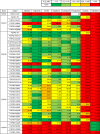
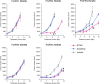
Various genetic alterations of the fibroblast growth factor receptor (FGFR) family have been detected across a wide range of cancers. However, inhibition of FGFR signaling by kinase inhibitors demonstrated limited clinical effectiveness. Herein, we evaluated the transforming activity and sensitivity of 160 nonsynonymous FGFR mutations and ten fusion genes to seven FGFR tyrosine kinase inhibitors (TKI) using the mixed-all-nominated-in-one (MANO) method, a high-throughput functional assay. The oncogenicity of 71 mutants was newly discovered in this study. The FGFR TKIs showed anti-proliferative activities against the wild-type FGFRs and their fusions, while several hotspot mutants were relatively resistant to those TKIs. The drug sensitivities assessed with the MANO method were well concordant with those evaluated using in vitro and in vivo assays. Comprehensive analysis of published FGFR structures revealed a possible mechanism through which oncogenic FGFR mutations reduce sensitivity to TKIs. It was further revealed that recurrent compound mutations within FGFRs affect the transforming potential and TKI-sensitivity of corresponding kinases. In conclusion, our study suggests the importance of selecting suitable inhibitors against individual FGFR variants. Moreover, it reveals the necessity to develop next-generation FGFR inhibitors, which are effective against all oncogenic FGFR variants.
© 2021. The Author(s).
Conflict of interest statement
S.K. reports research funding from Eisai Co., Ltd. The remaining authors declare no competing interests.
Figures


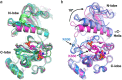
References
Grants and funding
LinkOut - more resources
Full Text Sources

