Bioinformatic Analysis of the Campylobacter jejuni Type VI Secretion System and Effector Prediction
- PMID: 34276628
- PMCID: PMC8285248
- DOI: 10.3389/fmicb.2021.694824
Bioinformatic Analysis of the Campylobacter jejuni Type VI Secretion System and Effector Prediction
Erratum in
-
Corrigendum: Bioinformatic Analysis of the Campylobacter jejuni Type VI Secretion System and Effector Prediction.Front Microbiol. 2021 Nov 16;12:793252. doi: 10.3389/fmicb.2021.793252. eCollection 2021. Front Microbiol. 2021. PMID: 34867930 Free PMC article.
Abstract
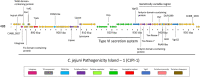
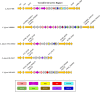
The Type VI Secretion System (T6SS) has important roles relating to bacterial antagonism, subversion of host cells, and niche colonisation. Campylobacter jejuni is one of the leading bacterial causes of human gastroenteritis worldwide and is a commensal coloniser of birds. Although recently discovered, the T6SS biological functions and identities of its effectors are still poorly defined in C. jejuni. Here, we perform a comprehensive bioinformatic analysis of the C. jejuni T6SS by investigating the prevalence and genetic architecture of the T6SS in 513 publicly available genomes using C. jejuni 488 strain as reference. A unique and conserved T6SS cluster associated with the Campylobacter jejuni Integrated Element 3 (CJIE3) was identified in the genomes of 117 strains. Analyses of the T6SS-positive 488 strain against the T6SS-negative C. jejuni RM1221 strain and the T6SS-positive plasmid pCJDM202 carried by C. jejuni WP2-202 strain defined the "T6SS-containing CJIE3" as a pathogenicity island, thus renamed as Campylobacter jejuni Pathogenicity Island-1 (CJPI-1). Analysis of CJPI-1 revealed two canonical VgrG homologues, CJ488_0978 and CJ488_0998, harbouring distinct C-termini in a genetically variable region downstream of the T6SS operon. CJPI-1 was also found to carry a putative DinJ-YafQ Type II toxin-antitoxin (TA) module, conserved across pCJDM202 and the genomic island CJIE3, as well as several open reading frames functionally predicted to encode for nucleases, lipases, and peptidoglycan hydrolases. This comprehensive in silico study provides a framework for experimental characterisation of T6SS-related effectors and TA modules in C. jejuni.
Keywords: Campylobacter jejuni; T6SS effectors; T6SS immunity proteins; Type VI Secretion System; pathogenicity island; toxin-antitoxin.
Copyright © 2021 Robinson, Liaw, Omole, Xia, van Vliet, Corcionivoschi, Hachani and Gundogdu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed October 5, 2020).
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

