Long Non-coding RNAs: Mechanisms, Experimental, and Computational Approaches in Identification, Characterization, and Their Biomarker Potential in Cancer
- PMID: 34276764
- PMCID: PMC8281131
- DOI: 10.3389/fgene.2021.649619
Long Non-coding RNAs: Mechanisms, Experimental, and Computational Approaches in Identification, Characterization, and Their Biomarker Potential in Cancer
Abstract
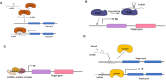
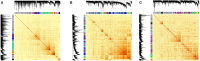
Long non-coding RNAs are diverse class of non-coding RNA molecules >200 base pairs of length having various functions like gene regulation, dosage compensation, epigenetic regulation. Dysregulation and genomic variations of several lncRNAs have been implicated in several diseases. Their tissue and developmental specific expression are contributing factors for them to be viable indicators of physiological states of the cells. Here we present an comprehensive review the molecular mechanisms and functions, state of the art experimental and computational pipelines and challenges involved in the identification and functional annotation of lncRNAs and their prospects as biomarkers. We also illustrate the application of co-expression networks on the TCGA-LIHC dataset for putative functional predictions of lncRNAs having a therapeutic potential in Hepatocellular carcinoma (HCC).
Keywords: biomarker; cancer; lncRNAs; mechanisms; methods.
Copyright © 2021 Chowdhary, Satagopam and Schneider.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Amodio N., Stamato M. A., Juli G., Morelli E., Fulciniti M., Manzoni M., et al. . (2018). Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. 32, 1948–1957. 10.1038/s41375-018-0067-3 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

