Identification of Atrial Fibrillation-Associated Genes ERBB2 and MYPN Using Genome-Wide Association and Transcriptome Expression Profile Data on Left-Right Atrial Appendages
- PMID: 34276800
- PMCID: PMC8278573
- DOI: 10.3389/fgene.2021.696591
Identification of Atrial Fibrillation-Associated Genes ERBB2 and MYPN Using Genome-Wide Association and Transcriptome Expression Profile Data on Left-Right Atrial Appendages
Abstract
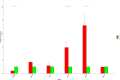
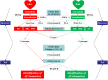
More reliable methods are needed to uncover novel biomarkers associated with atrial fibrillation (AF). Our objective is to identify significant network modules and newly AF-associated genes by integrative genetic analysis approaches. The single nucleotide polymorphisms with nominal relevance significance from the AF-associated genome-wide association study (GWAS) data were converted into the GWAS discovery set using ProxyGeneLD, followed by merging with significant network modules constructed by weighted gene coexpression network analysis (WGCNA) from one expression profile data set, composed of left and right atrial appendages (LAA and RAA). In LAA, two distinct network modules were identified (blue: p = 0.0076; yellow: p = 0.023). Five AF-associated biomarkers were identified (ERBB2, HERC4, MYH7, MYPN, and PBXIP1), combined with the GWAS test set. In RAA, three distinct network modules were identified and only one AF-associated gene LOXL1 was determined. Using human LAA tissues by real-time quantitative polymerase chain reaction, the differentially expressive results of ERBB2, MYH7, and MYPN were observed (p < 0.05). This study first demonstrated the feasibility of fusing GWAS with expression profile data by ProxyGeneLD and WGCNA to explore AF-associated genes. In particular, two newly identified genes ERBB2 and MYPN via this approach contribute to further understanding the occurrence and development of AF, thereby offering preliminary data for subsequent studies.
Keywords: atrial fibrillation; genome-wide association study; single nucleotide polymorphism; systems biology; transcriptome.
Copyright © 2021 Meng, Nie, Wang, Fan, Zhao and Yuan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Acciarresi M., Paciaroni M., Agnelli G., Falocci N., Caso V., Becattini C., et al. (2017). Prestroke CHA2DS2-VASc score and severity of acute stroke in patients with atrial fibrillation: findings from RAF study. J. Stroke Cerebrovasc. Dis. 26 1363–1368. 10.1016/j.jstrokecerebrovasdis.2017.02.011 - DOI - PubMed
-
- Barrios V., Calderon A., Escobar C., de la Figuera M. Primary Care Group in the Clinical Cardiology Section of the Spanish Society of Cardiology (2012). Patients with atrial fibrillation in a primary care setting: Val-FAAP study. Rev. Esp. Cardiol. 65 47–53. 10.1016/j.recesp.2011.08.008 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

