Type 1 conventional dendritic cells and interferons are required for spontaneous CD4+ and CD8+ T-cell protective responses to breast cancer
- PMID: 34277006
- PMCID: PMC8279130
- DOI: 10.1002/cti2.1305
Type 1 conventional dendritic cells and interferons are required for spontaneous CD4+ and CD8+ T-cell protective responses to breast cancer
Abstract
Objectives: To better understand how immune responses may be harnessed against breast cancer, we investigated which immune cell types and signalling pathways are required for spontaneous control of a mouse model of mammary adenocarcinoma.
Methods: The NOP23 mammary adenocarcinoma cell line expressing epitopes derived from the ovalbumin model antigen is spontaneously controlled when orthotopically engrafted in syngeneic C57BL/6 mice. We combined this breast cancer model with antibody-mediated depletion of lymphocytes and with mutant mice affected in interferon (IFN) or type 1 conventional dendritic cell (cDC1) responses. We monitored tumor growth and immune infiltration including the activation of cognate ovalbumin-specific T cells.
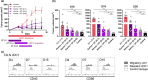
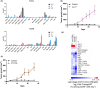
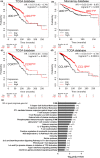
Results: Breast cancer immunosurveillance required cDC1, NK/NK T cells, conventional CD4+ T cells and CD8+ cytotoxic T lymphocytes (CTLs). cDC1 were required constitutively, but especially during T-cell priming. In tumors, cDC1 were interacting simultaneously with CD4+ T cells and tumor-specific CTLs. cDC1 expression of the XCR1 chemokine receptor and of the T-cell-attracting or T-cell-activating cytokines CXCL9, IL-12 and IL-15 was dispensable for tumor rejection, whereas IFN responses were necessary, including cDC1-intrinsic signalling by STAT1 and IFN-γ but not type I IFN (IFN-I). cDC1 and IFNs promoted CD4+ and CD8+ T-cell infiltration, terminal differentiation and effector functions. In breast cancer patients, high intratumor expression of genes specific to cDC1, CTLs, CD4+ T cells or IFN responses is associated with a better prognosis.
Conclusion: Interferons and cDC1 are critical for breast cancer immunosurveillance. IFN-γ plays a prominent role over IFN-I in licensing cDC1 for efficient T-cell activation.
Keywords: CD4+ T cells; CD8+ T cells; IFN‐γ; breast cancer; cDC1; cancer immunosurveillance; interferons.
© 2021 The Authors. Clinical & Translational Immunology published by John Wiley & Sons Australia, Ltd on behalf of Australian and New Zealand Society for Immunology, Inc.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
