Stimuli Responsive, Programmable DNA Nanodevices for Biomedical Applications
- PMID: 34277571
- PMCID: PMC8278982
- DOI: 10.3389/fchem.2021.704234
Stimuli Responsive, Programmable DNA Nanodevices for Biomedical Applications
Abstract
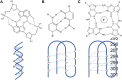
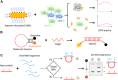
Of the multiple areas of applications of DNA nanotechnology, stimuli-responsive nanodevices have emerged as an elite branch of research owing to the advantages of molecular programmability of DNA structures and stimuli-responsiveness of motifs and DNA itself. These classes of devices present multiples areas to explore for basic and applied science using dynamic DNA nanotechnology. Herein, we take the stake in the recent progress of this fast-growing sub-area of DNA nanotechnology. We discuss different stimuli, motifs, scaffolds, and mechanisms of stimuli-responsive behaviours of DNA nanodevices with appropriate examples. Similarly, we present a multitude of biological applications that have been explored using DNA nanodevices, such as biosensing, in vivo pH-mapping, drug delivery, and therapy. We conclude by discussing the challenges and opportunities as well as future prospects of this emerging research area within DNA nanotechnology.
Keywords: DNA nanotechnology; biomedical applications; biosensing; stimulus responsive devices; therapeutics.
Copyright © 2021 Singh, Morya, Datta, Ghoroi and Bhatia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
LinkOut - more resources
Full Text Sources

