Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness
- PMID: 34279219
- PMCID: PMC8321553
- DOI: 10.7554/eLife.66277
Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness
Abstract
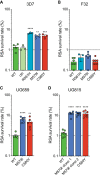
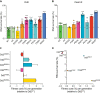
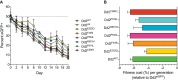
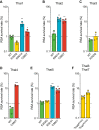
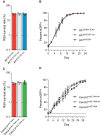
The emergence of mutant K13-mediated artemisinin (ART) resistance in Plasmodium falciparum malaria parasites has led to widespread treatment failures across Southeast Asia. In Africa, K13-propeller genotyping confirms the emergence of the R561H mutation in Rwanda and highlights the continuing dominance of wild-type K13 elsewhere. Using gene editing, we show that R561H, along with C580Y and M579I, confer elevated in vitro ART resistance in some African strains, contrasting with minimal changes in ART susceptibility in others. C580Y and M579I cause substantial fitness costs, which may slow their dissemination in high-transmission settings, in contrast with R561H that in African 3D7 parasites is fitness neutral. In Cambodia, K13 genotyping highlights the increasing spatio-temporal dominance of C580Y. Editing multiple K13 mutations into a panel of Southeast Asian strains reveals that only the R561H variant yields ART resistance comparable to C580Y. In Asian Dd2 parasites C580Y shows no fitness cost, in contrast with most other K13 mutations tested, including R561H. Editing of point mutations in ferredoxin or mdr2, earlier associated with resistance, has no impact on ART susceptibility or parasite fitness. These data underline the complex interplay between K13 mutations, parasite survival, growth and genetic background in contributing to the spread of ART resistance.
Keywords: CRISPR/Cas9; P. falciparum; Plasmodium falciparum; artemisinin resistance; epidemiology; fitness; global health; infectious disease; malaria; microbiology; ring-stage survival.
© 2021, Stokes et al.
Conflict of interest statement
BS, SD, KR, SM, JS, NG, ID, KS, JB, KW, JS, TY, LR, EL, FA, CC, IS, AG, RN, CN, AK, AU, SS, OK, MN, MW, RL, FN, TA, PR, DM, DF No competing interests declared
Figures




References
-
- Adams T, Ennuson NAA, Quashie NB, Futagbi G, Matrevi S, Hagan OCK, Abuaku B, Koram KA, Duah NO. Prevalence of Plasmodium falciparum delayed clearance associated polymorphisms in adaptor protein complex 2 mu subunit (pfap2mu) and ubiquitin specific protease 1 (pfubp1) genes in Ghanaian isolates. Parasites & Vectors. 2018;11:175. doi: 10.1186/s13071-018-2762-3. - DOI - PMC - PubMed
-
- Amambua-Ngwa A, Amenga-Etego L, Kamau E, Amato R, Ghansah A, Golassa L, Randrianarivelojosia M, Ishengoma D, Apinjoh T, Maïga-Ascofaré O, Andagalu B, Yavo W, Bouyou-Akotet M, Kolapo O, Mane K, Worwui A, Jeffries D, Simpson V, D'Alessandro U, Kwiatkowski D, Djimde AA. Major subpopulations of Plasmodium falciparum in sub-Saharan Africa. Science. 2019;365:813–816. doi: 10.1126/science.aav5427. - DOI - PubMed
-
- Amato R, Pearson RD, Almagro-Garcia J, Amaratunga C, Lim P, Suon S, Sreng S, Drury E, Stalker J, Miotto O, Fairhurst RM, Kwiatkowski DP. Origins of the current outbreak of multidrug-resistant malaria in Southeast Asia: a retrospective genetic study. The Lancet Infectious Diseases. 2018;18:337–345. doi: 10.1016/S1473-3099(18)30068-9. - DOI - PMC - PubMed
-
- Anderson TJ, Nair S, McDew-White M, Cheeseman IH, Nkhoma S, Bilgic F, McGready R, Ashley E, Pyae Phyo A, White NJ, Nosten F. Population parameters underlying an ongoing soft sweep in Southeast Asian malaria parasites. Molecular Biology and Evolution. 2017;34:131–144. doi: 10.1093/molbev/msw228. - DOI - PMC - PubMed
-
- Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

