Whole-genome sequencing of single circulating tumor cells from neuroendocrine neoplasms
- PMID: 34280125
- PMCID: PMC8428071
- DOI: 10.1530/ERC-21-0179
Whole-genome sequencing of single circulating tumor cells from neuroendocrine neoplasms
Abstract
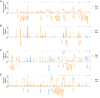
Single-cell profiling of circulating tumor cells (CTCs) as part of a minimally invasive liquid biopsy presents an opportunity to characterize and monitor tumor heterogeneity and evolution in individual patients. In this study, we aimed to compare single-cell copy number variation (CNV) data with tissue and define the degree of intra- and inter-patient genomic heterogeneity. We performed next-generation sequencing (NGS) whole-genome CNV analysis of 125 single CTCs derived from seven patients with neuroendocrine neoplasms (NEN) alongside matched white blood cells (WBC), formalin-fixed paraffin-embedded (FFPE), and fresh frozen (FF) samples. CTC CNV profiling demonstrated recurrent chromosomal alterations in previously reported NEN copy number hotspots, including the prognostically relevant loss of chromosome 18. Unsupervised hierarchical clustering revealed CTCs with distinct clonal lineages as well as significant intra- and inter-patient genomic heterogeneity, including subclonal alterations not detectable by bulk analysis and previously unreported in NEN. Notably, we also demonstrated the presence of genomically distinct CTCs according to the enrichment strategy utilized (EpCAM-dependent vs size-based). This work has significant implications for the identification of therapeutic targets, tracking of evolutionary change, and the implementation of CTC-biomarkers in cancer.
Keywords: circulating tumor cells; copy number variation; neuroendocrine tumors; single cell; whole-genome sequencing.
Figures





References
-
- Bolognesi C, Forcato C, Buson G, Fontana F, Mangano C, Doffini A, Sero V, Lanzellotto R, Signorini G, Calanca Aet al.2016Digital sorting of pure cell populations enables unambiguous genetic analysis of heterogeneous formalin-fixed paraffin-embedded tumors by next generation sequencing. Scientific Reports 620944. (10.1038/srep20944) - DOI - PMC - PubMed
-
- Bosman FT, Carneiro F, Hruban RH, Theise ND.2010WHO Classification of Tumours. IARC.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

