Molecular Characterization of TRPA Subfamily Genes and Function in Temperature Preference in Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae)
- PMID: 34281211
- PMCID: PMC8268038
- DOI: 10.3390/ijms22137157
Molecular Characterization of TRPA Subfamily Genes and Function in Temperature Preference in Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae)
Abstract
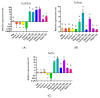
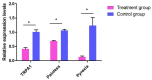
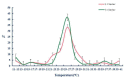
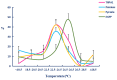
To reveal the mechanism of temperature preference in Tuta absoluta, one of the top 20 plant pests in the world, we cloned and identified TaTRPA1, TaPain, and TaPyx genes by RACE and bioinformatic analysis, and clarified their expression profiles during different development stages using real-time PCR, and revealed their function in preference temperature by RNAi. The full-length cDNA of TaPain was 3136 bp, with a 2865-bp open reading frame encoding a 259.89-kDa protein; and the partial length cDNA of TaPyx was 2326-bp, with a 2025-bp open reading frame encoding a 193.16-kDa protein. In addition, the expression of TaTRPA1 and TaPyx was significantly lower in larvae than other stages, and it was significantly higher in pupae and newly emerging males for TaPain. After feeding target double-stranded RNA (dsRNA), the preferred temperature decreased 2 °C more than the control group. In conclusion, the results firstly indicated the molecular characterization of TRPA subfamily genes and their key role in temperature perception in T. absoluta, and the study will help us to understand the temperature-sensing mechanism in the pest, and will provide some basis for study of other Lepidoptera insects' temperature preference. Moreover, it is of great significance in enriching the research progress of "thermos TRP".
Keywords: Painless; Pyrexia; RNA interference; TRPA1; Tuta absoluta; temperature preference.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Chromatin-Remodelling ATPases ISWI and BRM Are Essential for Reproduction in the Destructive Pest Tuta absoluta.Int J Mol Sci. 2022 Mar 17;23(6):3267. doi: 10.3390/ijms23063267. Int J Mol Sci. 2022. PMID: 35328688 Free PMC article.
-
DNA methylase 1 influences temperature responses and development in the invasive pest Tuta absoluta.Insect Mol Biol. 2024 Oct;33(5):503-515. doi: 10.1111/imb.12919. Epub 2024 May 29. Insect Mol Biol. 2024. PMID: 38808749
-
De novo transcriptome assembly and analysis to identify potential gene targets for RNAi-mediated control of the tomato leafminer (Tuta absoluta).BMC Genomics. 2015 Aug 26;16(1):635. doi: 10.1186/s12864-015-1841-5. BMC Genomics. 2015. PMID: 26306628 Free PMC article.
-
Effect of constant and fluctuating low temperature on the survival of Tuta absoluta pupae.Bull Entomol Res. 2024 Feb;114(1):1-7. doi: 10.1017/S0007485323000548. Epub 2023 Dec 15. Bull Entomol Res. 2024. PMID: 38098272
-
A gene silencing of V-ATPase subunit A interferes with survival and development of the tomato leafminer, Tuta absoluta.Arch Insect Biochem Physiol. 2021 Jan;106(1):e21753. doi: 10.1002/arch.21753. Epub 2020 Oct 30. Arch Insect Biochem Physiol. 2021. PMID: 33124713
Cited by
-
Avoidance of thiazoline compound depends on multiple sensory pathways mediated by TrpA1 and ORs in Drosophila.Front Mol Neurosci. 2023 Dec 22;16:1249715. doi: 10.3389/fnmol.2023.1249715. eCollection 2023. Front Mol Neurosci. 2023. PMID: 38188198 Free PMC article.
-
Chromatin-Remodelling ATPases ISWI and BRM Are Essential for Reproduction in the Destructive Pest Tuta absoluta.Int J Mol Sci. 2022 Mar 17;23(6):3267. doi: 10.3390/ijms23063267. Int J Mol Sci. 2022. PMID: 35328688 Free PMC article.
-
Mechanotransduction in Development: A Focus on Angiogenesis.Biology (Basel). 2025 Mar 27;14(4):346. doi: 10.3390/biology14040346. Biology (Basel). 2025. PMID: 40282211 Free PMC article. Review.
-
An effector of Phthorimaea absoluta oral secretions inhibits host plant defense.iScience. 2024 May 31;27(7):110154. doi: 10.1016/j.isci.2024.110154. eCollection 2024 Jul 19. iScience. 2024. PMID: 39050704 Free PMC article.
-
miR-252 targeting temperature receptor CcTRPM to mediate the transition from summer-form to winter-form of Cacopsylla chinensis.Elife. 2023 Nov 15;12:RP88744. doi: 10.7554/eLife.88744. Elife. 2023. PMID: 37965868 Free PMC article.
References
-
- Garcia M.F., Espul J.C. Bioecology of the tomato moth (Scrobipalpula absoluta) in Mendoza, Argentine Republic. Rev. Investig. Agrop. 1982;17:135–146.
-
- Desneux N., Wajnberg E., Wyckhuys K.A.G., Burgio G., Arpaia S., Narváez-Vasquez C.A., González-Cabrera J., Ruescas D.C., Tabone E., Frandon J., et al. Biological invasion of European tomato crops by Tuta absoluta: Ecology, geographic expansion and prospects for biological control. J. Pest Sci. 2010;83:197–215. doi: 10.1007/s10340-010-0321-6. - DOI
-
- Brévault T., Sylla S., Diatte M., Bernadas G., Diarra K. Tuta absoluta Meyrick (Lepidoptera: Gelechiidae): A New Threat to Tomato Production in Sub-Saharan Africa. Afr. Entomol. 2014;22:441–444. doi: 10.4001/003.022.0202. - DOI
-
- Chen L.M., Li X.W., He T.J., Li P.J., Liu Y., Zhou S.X., Wu Q.C., Chen T.T., Lu Y.B., Hou Y.M. Comparative biochemical and transcriptome analyses in tomato and eggplant reveal their differential responses to Tuta absoluta infestation. Genomics. 2021;113:2108–2121. doi: 10.1016/j.ygeno.2021.05.002. - DOI - PubMed
-
- Zhang G.F., Ma D.Y., Wang Y.S.H., Gao Y.H., Liu W.X., Zhang R., Fu W.J., Xian X.Q., Wang J., Kuang M., et al. First report of the South American tomato leafminer, Tuta absoluta (Meyrick), in China. J. Integr. Agr. 2020;19:1912–1917. doi: 10.1016/S2095-3119(20)63165-3. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

