Evolutionary Tracking of SARS-CoV-2 Genetic Variants Highlights an Intricate Balance of Stabilizing and Destabilizing Mutations
- PMID: 34281387
- PMCID: PMC8406184
- DOI: 10.1128/mBio.01188-21
Evolutionary Tracking of SARS-CoV-2 Genetic Variants Highlights an Intricate Balance of Stabilizing and Destabilizing Mutations
Abstract
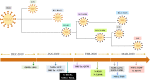
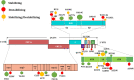
The currently ongoing COVID-19 pandemic caused by SARS-CoV-2 has accounted for millions of infections and deaths across the globe. Genome sequences of SARS-CoV-2 are being published daily in public databases and the availability of these genome data sets has allowed unprecedented access to the mutational patterns of SARS-CoV-2 evolution. We made use of the same genomic information for conducting phylogenetic analysis and identifying lineage-specific mutations. The catalogued lineage-defining mutations were analyzed for their stabilizing or destabilizing impact on viral proteins. We recorded persistence of D614G, S477N, A222V, and V1176F variants and a global expansion of the PANGOLIN variant B.1. In addition, a retention of Q57H (B.1.X), R203K/G204R (B.1.1.X), T85I (B.1.2-B.1.3), G15S+T428I (C.X), and I120F (D.X) variations was observed. Overall, we recorded a striking balance between stabilizing and destabilizing mutations, therefore leading to well-maintained protein structures. With selection pressures in the form of newly developed vaccines and therapeutics to mount in the coming months, the task of mapping viral mutations and recording their impact on key viral proteins should be crucial to preemptively catch any escape mechanism for which SARS-CoV-2 may evolve. IMPORTANCE Since its initial isolation in Wuhan, China, large numbers of SARS-CoV-2 genome sequences have been shared in publicly accessible repositories, thus enabling scientists to do detailed evolutionary analysis. We investigated the evolutionarily associated mutational diversity overlaid on the major phylogenetic lineages circulating globally, using 513 representative genomes. We detailed the phylogenetic persistence of key variants facilitating global expansion of the PANGOLIN variant B.1, including the recent, fast-expanding, B.1.1.7 lineage. The stabilizing or destabilizing impact of the catalogued lineage-defining mutations on viral proteins indicates their possible involvement in balancing the protein function and structure. A clear understanding of this mutational profile is of high clinical significance to catch any vaccine escape mechanism, as the same proteins make crucial components of vaccines that have recently been approved or are in development. In this vein, our study provides an imperative framework and baseline data upon which further analysis could be built as newer variants of SARS-CoV-2 continue to appear.
Keywords: SARS-CoV-2; evolution; mutation; stability; vaccine.
Figures

Similar articles
-
Evolution, Mode of Transmission, and Mutational Landscape of Newly Emerging SARS-CoV-2 Variants.mBio. 2021 Aug 31;12(4):e0114021. doi: 10.1128/mBio.01140-21. Epub 2021 Aug 31. mBio. 2021. PMID: 34465019 Free PMC article.
-
The extent of molecular variation in novel SARS-CoV-2 after the six-month global spread.Infect Genet Evol. 2021 Jul;91:104800. doi: 10.1016/j.meegid.2021.104800. Epub 2021 Mar 5. Infect Genet Evol. 2021. PMID: 33677109 Free PMC article.
-
Global variation in SARS-CoV-2 proteome and its implication in pre-lockdown emergence and dissemination of 5 dominant SARS-CoV-2 clades.Infect Genet Evol. 2021 Sep;93:104973. doi: 10.1016/j.meegid.2021.104973. Epub 2021 Jun 18. Infect Genet Evol. 2021. PMID: 34147651 Free PMC article.
-
The variants question: What is the problem?J Med Virol. 2021 Dec;93(12):6479-6485. doi: 10.1002/jmv.27196. Epub 2021 Jul 28. J Med Virol. 2021. PMID: 34255352 Free PMC article. Review.
-
Implication of SARS-CoV-2 Immune Escape Spike Variants on Secondary and Vaccine Breakthrough Infections.Front Immunol. 2021 Nov 3;12:742167. doi: 10.3389/fimmu.2021.742167. eCollection 2021. Front Immunol. 2021. PMID: 34804022 Free PMC article. Review.
Cited by
-
Delta variant (B.1.617.2) of SARS-CoV-2: Mutations, impact, challenges and possible solutions.Hum Vaccin Immunother. 2022 Nov 30;18(5):2068883. doi: 10.1080/21645515.2022.2068883. Epub 2022 May 4. Hum Vaccin Immunother. 2022. PMID: 35507895 Free PMC article. Review.
-
Global Analysis of Tracking the Evolution of SARS-CoV-2 Variants.Vaccines (Basel). 2023 Dec 4;11(12):1812. doi: 10.3390/vaccines11121812. Vaccines (Basel). 2023. PMID: 38140216 Free PMC article.
-
Epimaps of the SARS-CoV-2 Receptor-Binding Domain Mutational Landscape: Insights into Protein Stability, Epitope Prediction, and Antibody Binding.Biomolecules. 2025 Feb 18;15(2):301. doi: 10.3390/biom15020301. Biomolecules. 2025. PMID: 40001604 Free PMC article.
-
Neutralising antibody escape of SARS-CoV-2 spike protein: Risk assessment for antibody-based Covid-19 therapeutics and vaccines.Rev Med Virol. 2021 Nov;31(6):e2231. doi: 10.1002/rmv.2231. Epub 2021 Mar 16. Rev Med Virol. 2021. PMID: 33724631 Free PMC article. Review.
-
Unique SARS-CoV-2 Variants, Tourism Metrics, and B.1.2 Emergence in Early COVID-19 Pandemic: A Correlation Analysis in South Dakota.Int J Environ Res Public Health. 2023 Sep 13;20(18):6748. doi: 10.3390/ijerph20186748. Int J Environ Res Public Health. 2023. PMID: 37754608 Free PMC article.
References
-
- World Health Organization. 2020. Coronavirus disease 2019 (COVID-19): situation report. World Health Organization, Geneva, Switzerland.
-
- Worldometer. 2020. COVID-19 Coronavirus. https://www.worldometers.info/coronavirus/. Accessed 27 December 2020.
-
- World Health Organization. 2020. Rolling updates on coronavirus disease (COVID19). https://www.who.int/emergencies/diseases/novel-coronavirus2019/events-as.... Accessed 18 May 2020.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

