Leucoefdin a potential inhibitor against SARS CoV-2 Mpro
- PMID: 34281489
- PMCID: PMC7309301
- DOI: 10.1080/07391102.2020.1777903
Leucoefdin a potential inhibitor against SARS CoV-2 Mpro
Abstract
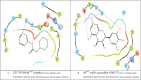
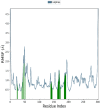
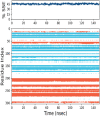
Leucoefdin an important constituent of various fruits such as banana, raspberry, etc. was explored to target MPro protease of SARS Co-V 2. Ligand was found to bind at active site of MPro with large negative binding energies in molecular docking and simulation study. The docking results showed that Leucoefdin interacted with the MPro by forming hydrogen bonds, at Leu 141, His163, His 164, and Glu 166. Other non-bonded interactions were seen at Met49, Pro52, Tyr54, Phe140, Leu141, Cys145 and Met165. Results of Leucoefdin was in coherence with the recently reported MPro protease-inhibitor complex. It even displayed better binding energies (kcal/mol) in HTVS (-6.28), SP (-7.28), XP (-9.29) and MMGBSA (-44.71) as compared to the reference ligand [HTVS (-4.87), SP (-6.79), XP (-5.75) and MMGBSA (-47.76)]. Leucoefdin-MPro complex on molecular dynamic simulation showed initial fluctuations in RMSD plot for a certain period and attained equilibrium which remained stable during entire simulation for 150 ns. RMSF of protein showed less secondary structure fluctuations and a greater number of H-bond formation with Leucoefdin during 150 ns simulation. Post simulation MMGBSA analysis showed binding energy of -45.98 Kcal/mol. These findings indicated the potential of Leucoefdin as lead compound in R&D for drug discovery and development against SARS CoV-2.Communicated by Ramaswamy H. Sarma.
Keywords: Leucoefdin; MPro; SARS CoV-2; docking; simulation.
Figures



Similar articles
-
Molecular insights to the binding interactions of APNS containing HIV-protease inhibitors against SARS-CoV-2 Mpro: an in silico approach towards drug repurposing.J Biomol Struct Dyn. 2023 Jun;41(9):3900-3913. doi: 10.1080/07391102.2022.2059008. Epub 2022 Apr 7. J Biomol Struct Dyn. 2023. PMID: 35388744
-
Insight into crystal structures and identification of potential styrylthieno[2,3-b]pyridine-2-carboxamidederivatives against COVID-19 Mpro through structure-guided modeling and simulation approach.J Biomol Struct Dyn. 2024 May;42(8):4325-4343. doi: 10.1080/07391102.2023.2220799. Epub 2023 Jun 15. J Biomol Struct Dyn. 2024. PMID: 37318002
-
Virtual screening of natural products inspired in-house library to discover potential lead molecules against the SARS-CoV-2 main protease.J Biomol Struct Dyn. 2023 Mar;41(5):2033-2045. doi: 10.1080/07391102.2022.2027271. Epub 2022 Jan 19. J Biomol Struct Dyn. 2023. PMID: 35043750
-
Identification of Natural Inhibitors Against SARS-CoV-2 Drugable Targets Using Molecular Docking, Molecular Dynamics Simulation, and MM-PBSA Approach.Front Cell Infect Microbiol. 2021 Aug 12;11:730288. doi: 10.3389/fcimb.2021.730288. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34458164 Free PMC article.
-
In silico identification of potential inhibitors of key SARS-CoV-2 3CL hydrolase (Mpro) via molecular docking, MMGBSA predictive binding energy calculations, and molecular dynamics simulation.PLoS One. 2020 Jul 24;15(7):e0235030. doi: 10.1371/journal.pone.0235030. eCollection 2020. PLoS One. 2020. PMID: 32706783 Free PMC article.
Cited by
-
Structure-guided discovery of a novel BTK inhibitor inducing apoptosis and G1 phase arrest in tumor cells.Mol Divers. 2025 Aug 31. doi: 10.1007/s11030-025-11334-z. Online ahead of print. Mol Divers. 2025. PMID: 40886252
-
Computational analysis of dynamic allostery and control in the SARS-CoV-2 main protease.J R Soc Interface. 2021 Jan;18(174):20200591. doi: 10.1098/rsif.2020.0591. Epub 2021 Jan 6. J R Soc Interface. 2021. PMID: 33402024 Free PMC article.
-
Methodology-Centered Review of Molecular Modeling, Simulation, and Prediction of SARS-CoV-2.Chem Rev. 2022 Jul 13;122(13):11287-11368. doi: 10.1021/acs.chemrev.1c00965. Epub 2022 May 20. Chem Rev. 2022. PMID: 35594413 Free PMC article. Review.
-
Potentials of Antitussive Traditional Persian Functional Foods for COVID-19 Therapy†.Front Pharmacol. 2021 Jul 16;12:624006. doi: 10.3389/fphar.2021.624006. eCollection 2021. Front Pharmacol. 2021. PMID: 34335237 Free PMC article. Review.
-
Plants Metabolites: Possibility of Natural Therapeutics Against the COVID-19 Pandemic.Front Med (Lausanne). 2020 Aug 7;7:444. doi: 10.3389/fmed.2020.00444. eCollection 2020. Front Med (Lausanne). 2020. PMID: 32850918 Free PMC article. Review.
References
-
- Agostini M. L., Andres E. L., Sims A. C., Graham R. L., Sheahan T. P., Lu X., Smith E. C., Case J. B., Feng J. Y., Jordan R., Ray A. S., Cihlar T., Siegel D., Mackman R. L., Clarke M. O., Baric R. S., & Denison M. R. (2018). Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading Exoribonuclease. mBio, 9(2), 6 10.1128/mBio.00221-18 - DOI - PMC - PubMed
-
- Fearon D., Powell A. J., Douangamath A., Owen C. D., Wild C., Krojer T., Lukacik P., Strain-Damerell C. M., Walsh M. A., & von Delft F. (2020). PanDDA analysis group deposition—Crystal structure of COVID-19 main protease in complex with Z31792168. DOI: 10.2210/pdb5R84/pdb. - DOI
-
- Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shaw D. E., Shelley M., Perry J. K., Francis P., & Shenkin P. S. (2004). Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry, 47(7), 1739–1749. 10.1021/jm0306430 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
