The core autophagy machinery is not required for chloroplast singlet oxygen-mediated cell death in the Arabidopsis thaliana plastid ferrochelatase two mutant
- PMID: 34281507
- PMCID: PMC8290626
- DOI: 10.1186/s12870-021-03119-x
The core autophagy machinery is not required for chloroplast singlet oxygen-mediated cell death in the Arabidopsis thaliana plastid ferrochelatase two mutant
Abstract
Background: Chloroplasts respond to stress and changes in the environment by producing reactive oxygen species (ROS) that have specific signaling abilities. The ROS singlet oxygen (1O2) is unique in that it can signal to initiate cellular degradation including the selective degradation of damaged chloroplasts. This chloroplast quality control pathway can be monitored in the Arabidopsis thaliana mutant plastid ferrochelatase two (fc2) that conditionally accumulates chloroplast 1O2 under diurnal light cycling conditions leading to rapid chloroplast degradation and eventual cell death. The cellular machinery involved in such degradation, however, remains unknown. Recently, it was demonstrated that whole damaged chloroplasts can be transported to the central vacuole via a process requiring autophagosomes and core components of the autophagy machinery. The relationship between this process, referred to as chlorophagy, and the degradation of 1O2-stressed chloroplasts and cells has remained unexplored.
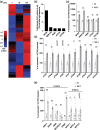
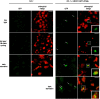
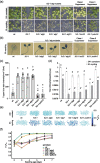
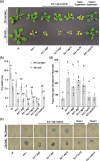
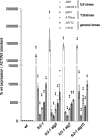
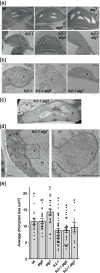
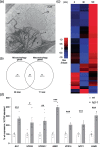
Results: To further understand 1O2-induced cellular degradation and determine what role autophagy may play, the expression of autophagy-related genes was monitored in 1O2-stressed fc2 seedlings and found to be induced. Although autophagosomes were present in fc2 cells, they did not associate with chloroplasts during 1O2 stress. Mutations affecting the core autophagy machinery (atg5, atg7, and atg10) were unable to suppress 1O2-induced cell death or chloroplast protrusion into the central vacuole, suggesting autophagosome formation is dispensable for such 1O2-mediated cellular degradation. However, both atg5 and atg7 led to specific defects in chloroplast ultrastructure and photosynthetic efficiencies, suggesting core autophagy machinery is involved in protecting chloroplasts from photo-oxidative damage. Finally, genes predicted to be involved in microautophagy were shown to be induced in stressed fc2 seedlings, indicating a possible role for an alternate form of autophagy in the dismantling of 1O2-damaged chloroplasts.
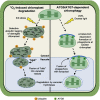
Conclusions: Our results support the hypothesis that 1O2-dependent cell death is independent from autophagosome formation, canonical autophagy, and chlorophagy. Furthermore, autophagosome-independent microautophagy may be involved in degrading 1O2-damaged chloroplasts. At the same time, canonical autophagy may still play a role in protecting chloroplasts from 1O2-induced photo-oxidative stress. Together, this suggests chloroplast function and degradation is a complex process utilizing multiple autophagy and degradation machineries, possibly depending on the type of stress or damage incurred.
Keywords: Abiotic stress; Autophagy; Cellular degradation; Chloroplast; Microautophagy; Oxidative stress; Photosynthesis; Reactive oxygen species; Signaling; Singlet oxygen.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

