SARS-CoV-2: from its discovery to genome structure, transcription, and replication
- PMID: 34281608
- PMCID: PMC8287290
- DOI: 10.1186/s13578-021-00643-z
SARS-CoV-2: from its discovery to genome structure, transcription, and replication
Abstract
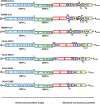
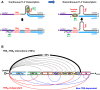
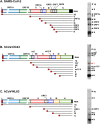
SARS-CoV-2 is an extremely contagious respiratory virus causing adult atypical pneumonia COVID-19 with severe acute respiratory syndrome (SARS). SARS-CoV-2 has a single-stranded, positive-sense RNA (+RNA) genome of ~ 29.9 kb and exhibits significant genetic shift from different isolates. After entering the susceptible cells expressing both ACE2 and TMPRSS2, the SARS-CoV-2 genome directly functions as an mRNA to translate two polyproteins from the ORF1a and ORF1b region, which are cleaved by two viral proteases into sixteen non-structural proteins (nsp1-16) to initiate viral genome replication and transcription. The SARS-CoV-2 genome also encodes four structural (S, E, M and N) and up to six accessory (3a, 6, 7a, 7b, 8, and 9b) proteins, but their translation requires newly synthesized individual subgenomic RNAs (sgRNA) in the infected cells. Synthesis of the full-length viral genomic RNA (gRNA) and sgRNAs are conducted inside double-membrane vesicles (DMVs) by the viral replication and transcription complex (RTC), which comprises nsp7, nsp8, nsp9, nsp12, nsp13 and a short RNA primer. To produce sgRNAs, RTC starts RNA synthesis from the highly structured gRNA 3' end and switches template at various transcription regulatory sequence (TRSB) sites along the gRNA body probably mediated by a long-distance RNA-RNA interaction. The TRS motif in the gRNA 5' leader (TRSL) is responsible for the RNA-RNA interaction with the TRSB upstream of each ORF and skipping of the viral genome in between them to produce individual sgRNAs. Abundance of individual sgRNAs and viral gRNA synthesized in the infected cells depend on the location and read-through efficiency of each TRSB. Although more studies are needed, the unprecedented COVID-19 pandemic has taught the world a painful lesson that is to invest and proactively prepare future emergence of other types of coronaviruses and any other possible biological horrors.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

