Sirtuin 6: linking longevity with genome and epigenome stability
- PMID: 34281779
- PMCID: PMC8903056
- DOI: 10.1016/j.tcb.2021.06.009
Sirtuin 6: linking longevity with genome and epigenome stability
Abstract
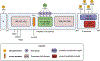
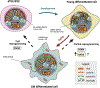
Sirtuin 6 (SIRT6) has been in the spotlight of aging research because progeroid phenotypes are associated with SIRT6 deficiency. SIRT6 has multiple molecular functions, including DNA repair and heterochromatin regulation, which position SIRT6 as a hub that regulates genome and epigenome stability. Genomic instability caused by persistent DNA damage and accumulating mutations, together with alterations in the epigenetic landscape and derepression of repetitive genetic elements, have emerged as mechanisms driving organismal aging. Enhanced levels of SIRT6 expression or activity provide avenues for rejuvenation strategies. This review focuses on the role of SIRT6 in the maintenance of genome and epigenome stability and its link to longevity. We propose a model where SIRT6 together with lamins control aging and rejuvenation by maintaining epigenetic silencing of repetitive elements.
Keywords: DNA damage repair; SIRT6; epigenome; healthspan; longevity; rejuvenation.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no conflicts of interest.
Figures
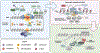
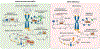
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

