Tradeoffs for a viral mutant with enhanced replication speed
- PMID: 34282021
- PMCID: PMC8325337
- DOI: 10.1073/pnas.2105288118
Tradeoffs for a viral mutant with enhanced replication speed
Abstract
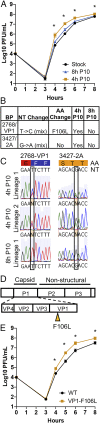
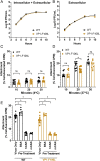
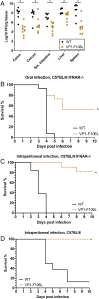
RNA viruses exist as genetically heterogeneous populations due to high mutation rates, and many of these mutations reduce fitness and/or replication speed. However, it is unknown whether mutations can increase replication speed of a virus already well adapted to replication in cultured cells. By sequentially passaging coxsackievirus B3 in cultured cells and collecting the very earliest progeny, we selected for increased replication speed. We found that a single mutation in a viral capsid protein, VP1-F106L, was sufficient for the fast-replication phenotype. Characterization of this mutant revealed quicker genome release during entry compared to wild-type virus, highlighting a previously unappreciated infection barrier. However, this mutation also reduced capsid stability in vitro and reduced replication and pathogenesis in mice. These results reveal a tradeoff between overall replication speed and fitness. Importantly, this approach-selecting for the earliest viral progeny-could be applied to a variety of viral systems and has the potential to reveal unanticipated inefficiencies in viral replication cycles.
Keywords: capsid; coxsackievirus; fitness; replication speed.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

