MX2-mediated innate immunity against HIV-1 is regulated by serine phosphorylation
- PMID: 34282309
- PMCID: PMC7611661
- DOI: 10.1038/s41564-021-00937-5
MX2-mediated innate immunity against HIV-1 is regulated by serine phosphorylation
Erratum in
-
Author Correction: MX2-mediated innate immunity against HIV-1 is regulated by serine phosphorylation.Nat Microbiol. 2021 Sep;6(9):1211. doi: 10.1038/s41564-021-00960-6. Nat Microbiol. 2021. PMID: 34389816 No abstract available.
Abstract
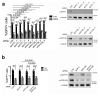
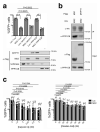
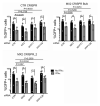
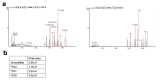
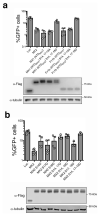
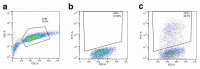
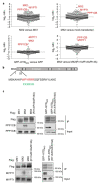
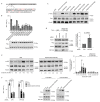
The antiviral cytokine interferon activates expression of interferon-stimulated genes to establish an antiviral state. Myxovirus resistance 2 (MX2, also known as MxB) is an interferon-stimulated gene that inhibits the nuclear import of HIV-1 and interacts with the viral capsid and cellular nuclear transport machinery. Here, we identified the myosin light chain phosphatase (MLCP) subunits myosin phosphatase target subunit 1 (MYPT1) and protein phosphatase 1 catalytic subunit-β (PPP1CB) as positively-acting regulators of MX2, interacting with its amino-terminal domain. We demonstrated that serine phosphorylation of the N-terminal domain at positions 14, 17 and 18 suppresses MX2 antiviral function, prevents interactions with the HIV-1 capsid and nuclear transport factors, and is reversed by MLCP. Notably, serine phosphorylation of the N-terminal domain also impedes MX2-mediated inhibition of nuclear import of cellular karyophilic cargo. We also found that interferon treatment reduces levels of phosphorylation at these serine residues and outline a homeostatic regulatory mechanism in which repression of MX2 by phosphorylation, together with MLCP-mediated dephosphorylation, balances the deleterious effects of MX2 on normal cell function with innate immunity against HIV-1.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Liu Z, et al. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe. 2013;14:398–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

