High-throughput and single-cell T cell receptor sequencing technologies
- PMID: 34282327
- PMCID: PMC9345561
- DOI: 10.1038/s41592-021-01201-8
High-throughput and single-cell T cell receptor sequencing technologies
Abstract
T cells express T cell receptors (TCRs) composed of somatically recombined TCRα and TCRβ chains, which mediate recognition of major histocompatibility complex (MHC)-antigen complexes and drive the antigen-specific adaptive immune response to pathogens and cancer. The TCR repertoire in each individual is highly diverse, which allows for recognition of a wide array of foreign antigens, but also presents a challenge in analyzing this response using conventional methods. Recent studies have developed high-throughput sequencing technologies to identify TCR sequences, analyze their antigen specificities using experimental and computational tools, and pair TCRs with transcriptional and epigenetic cell state phenotypes in single cells. In this Review, we highlight these technological advances and describe how they have been applied to discover fundamental insights into T cell-mediated immunity.
© 2021. Springer Nature America, Inc.
Figures

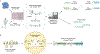
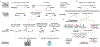
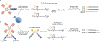
References
-
- Davis MM & Bjorkman PJ The T cell receptor genes and T cell recognition. Nature 334, 395–402 (1988). - PubMed
-
- Nikolich-Žugich J, Slifka MK & Messaoudi I The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol 4, 123–132 (2004). - PubMed
-
- Bradley P & Thomas PG Using T cell receptor repertoires to understand the principles of adaptive immune recognition. Annu. Rev. Immunol 37, 547–570 (2019). - PubMed
-
- Arstila TP et al. A direct estimate of the human αβ T cell receptor diversity. Science 286, 958–961 (1999). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

